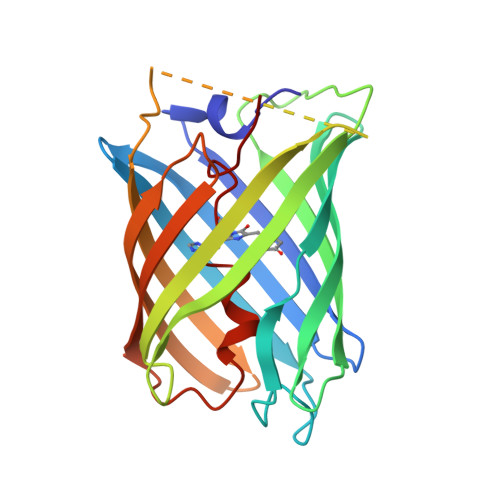
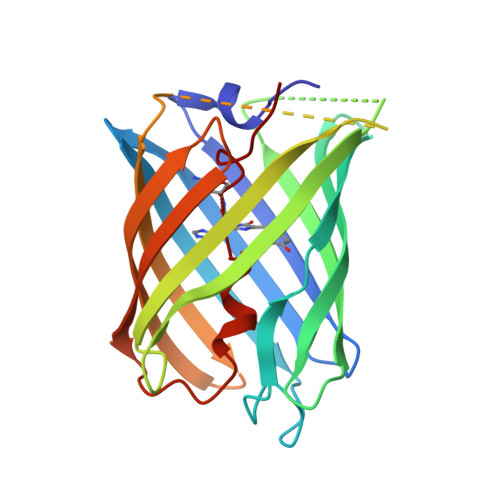
Photocleavable proteins that undergo fast and efficient dissociation.
Lu, X., Wen, Y., Zhang, S., Zhang, W., Chen, Y., Shen, Y., Lemieux, M.J., Campbell, R.E.(2021) Chem Sci 12: 9658-9672
- PubMed: 34349937
- DOI: https://doi.org/10.1039/d1sc01059j
- Primary Citation of Related Structures:
7DMX, 7DNA, 7DNB - PubMed Abstract:
Photocleavable molecules can enable the light-dependent modulation of biomolecular activities with high spatiotemporal precision. We have previously reported a photocleavable protein (PhoCl1) that, uniquely, is a fully genetically encoded photocleavable molecule that can be introduced into cells in the form of its corresponding gene to enable optogenetic control of biomolecular activities. However, the first generation PhoCl1 exhibited a relatively slow rate of dissociation, potentially limiting its utility. Here, we report the X-ray crystal structures of the PhoCl1 green state, red state, and cleaved empty barrel. Molecular dynamics (MD) simulations were performed to provide insight into the precise dissociation mechanism. Using structure-guided engineering and directed evolution, we have developed PhoCl2c with higher contrast ratio and PhoCl2f with faster dissociation. We characterized the performance of these new variants as purified proteins and in cultured cells. Our results demonstrate that PhoCl2 variants exhibit faster and more efficient dissociation, which should enable improved optogenetic manipulations of protein localization and protein-protein interactions in living cells.
- Department of Chemistry, University of Alberta Edmonton Alberta T6G 2G2 Canada robert.e.campbell@ualberta.ca.
Organizational Affiliation: