Non-canonical phosphorylation of Bmf by p38 MAPK promotes its apoptotic activity in anoikis.
Zhi, Z., Ouyang, Z., Ren, Y., Cheng, Y., Liu, P., Wen, Y., Shao, Y.(2022) Cell Death Differ 29: 323-336
- PubMed: 34462553
- DOI: https://doi.org/10.1038/s41418-021-00855-3
- Primary Citation of Related Structures:
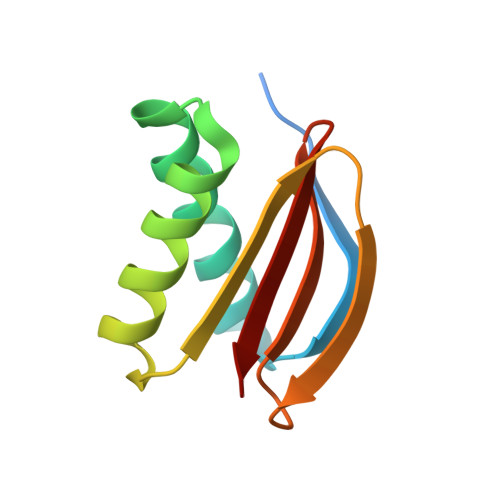
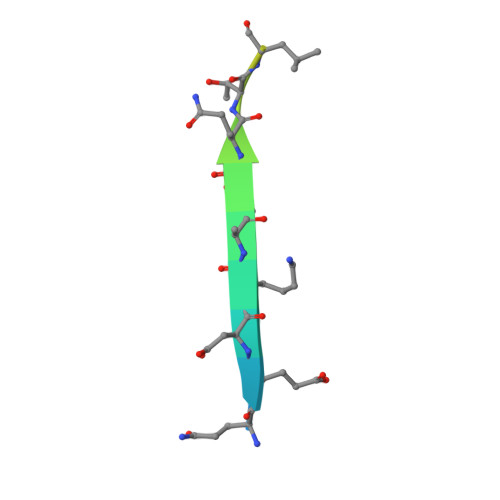
7CNU - PubMed Abstract:
Bmf contributes to the onset of anoikis by translocating from cytoskeleton to mitochondria when cells lose attachment to the extracellular matrix. However, the structural details of Bmf cytoskeleton tethering and the control of Bmf release upon loss of anchorage remained unknown. Here we showed that cell detachment induced rapid and sustained activation of p38 MAPK in mammary epithelial cell lines. Inhibition of p38 signaling or Bmf knockdown rescued anoikis. Activated p38 MAPK could directly phosphorylate Bmf at multiple sites including a non-proline-directed site threonine 72 (T72). Crystallographic studies revealed that Bmf T72 directly participated in DLC2 binding and its phosphorylation would block Bmf/DLC2 interaction through steric hindrance. Finally, we showed that phosphomimetic mutation of T72 enhanced Bmf apoptotic activity in vitro and in a knock-in mouse model. This work unraveled a novel regulatory mechanism of Bmf activity during anoikis and provided structural basis for Bmf cytoskeleton tethering and dissociation.
- Frontier Institute of Science and Technology, Xi'an Jiaotong University, Xi'an, China.
Organizational Affiliation: