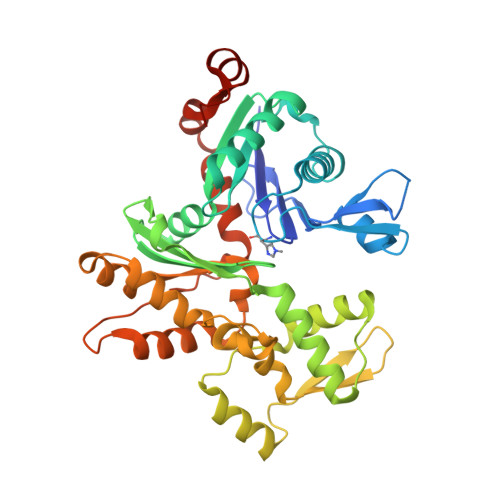
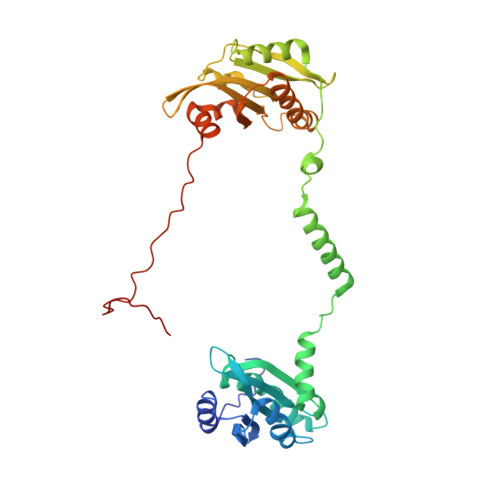
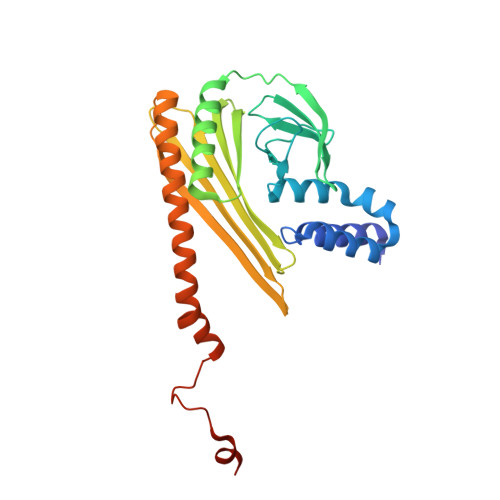
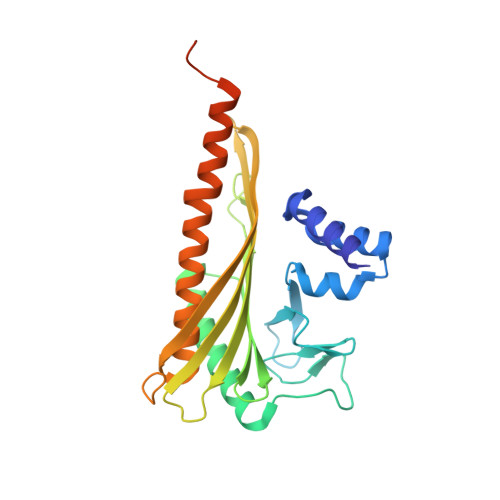
The structure of the actin filament uncapping complex mediated by twinfilin.
Mwangangi, D.M., Manser, E., Robinson, R.C.(2021) Sci Adv 7
- PubMed: 33571120
- DOI: https://doi.org/10.1126/sciadv.abd5271
- Primary Citation of Related Structures:
7CCC - PubMed Abstract:
Uncapping of actin filaments is essential for driving polymerization and depolymerization dynamics from capping protein-associated filaments; however, the mechanisms of uncapping leading to rapid disassembly are unknown. Here, we elucidated the x-ray crystal structure of the actin/twinfilin/capping protein complex to address the mechanisms of twinfilin uncapping of actin filaments. The twinfilin/capping protein complex binds to two G-actin subunits in an orientation that resembles the actin filament barbed end. This suggests an unanticipated mechanism by which twinfilin disrupts the stable capping of actin filaments by inducing a G-actin conformation in the two terminal actin subunits. Furthermore, twinfilin disorders critical actin-capping protein interactions, which will assist in the dissociation of capping protein, and may promote filament uncapping through a second mechanism involving V-1 competition for an actin-binding surface on capping protein. The extensive interactions with capping protein indicate that the evolutionary conserved role of twinfilin is to uncap actin filaments.
- Institute of Molecular and Cell Biology, A*STAR (Agency for Science, Technology and Research), Biopolis, Singapore 138673, Singapore.
Organizational Affiliation: