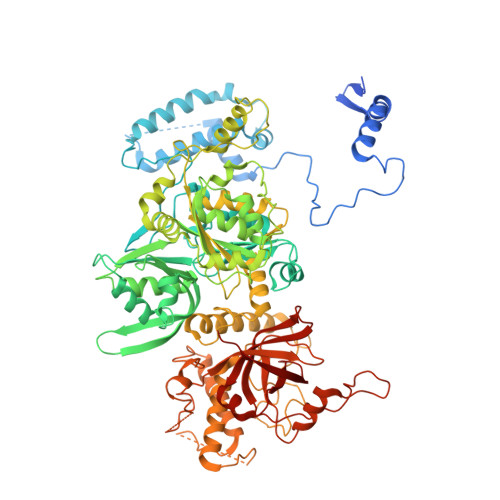
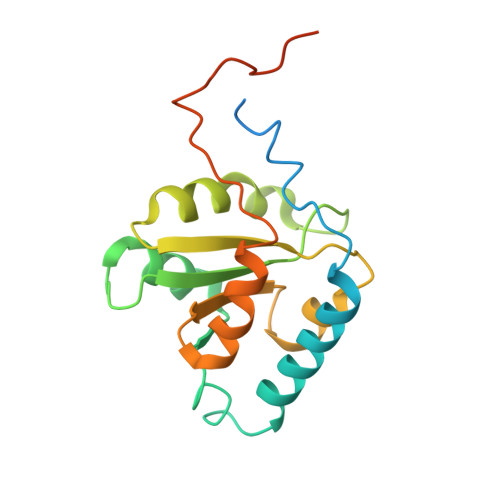
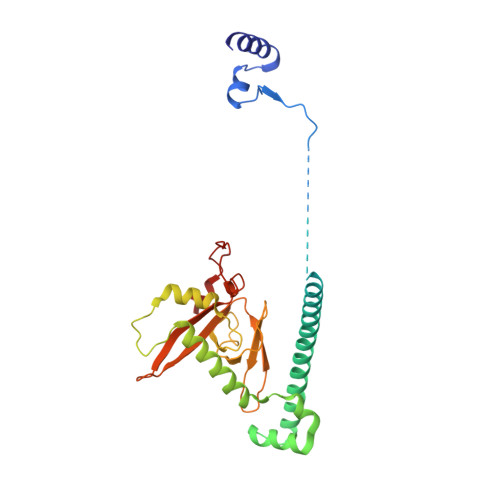
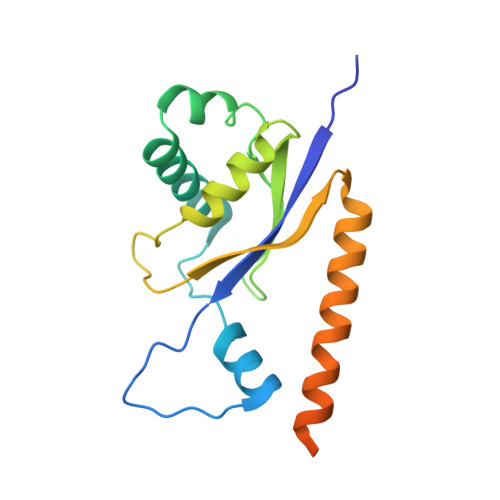
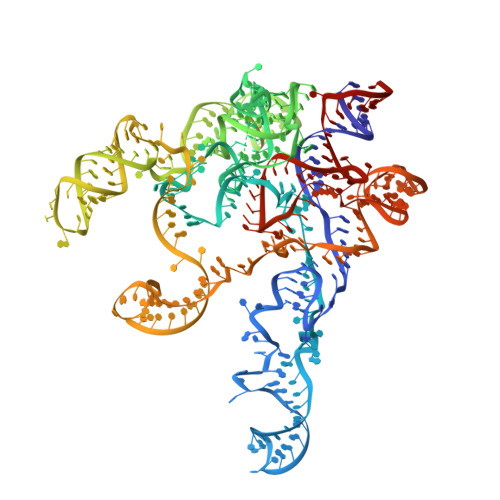
Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Lan, P., Zhou, B., Tan, M., Li, S., Cao, M., Wu, J., Lei, M.(2020) Science 369: 656-663
- PubMed: 32586950
- DOI: https://doi.org/10.1126/science.abc0149
- Primary Citation of Related Structures:
7C79, 7C7A - PubMed Abstract:
Ribonuclease (RNase) MRP is a conserved eukaryotic ribonucleoprotein complex that plays essential roles in precursor ribosomal RNA (pre-rRNA) processing and cell cycle regulation. In contrast to RNase P, which selectively cleaves transfer RNA-like substrates, it has remained a mystery how RNase MRP recognizes its diverse substrates. To address this question, we determined cryo-electron microscopy structures of Saccharomyces cerevisiae RNase MRP alone and in complex with a fragment of pre-rRNA. These structures and the results of biochemical studies reveal that coevolution of both protein and RNA subunits has transformed RNase MRP into a distinct ribonuclease that processes single-stranded RNAs by recognizing a short, loosely defined consensus sequence. This broad substrate specificity suggests that RNase MRP may have myriad yet unrecognized substrates that could play important roles in various cellular contexts.
- State Key Laboratory of Oncogenes and Related Genes, Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, China.
Organizational Affiliation: