Structural Insights into a Novel Esterase from the East Pacific Rise and Its Improved Thermostability by a Semirational Design.
Zhu, C., Chen, Y., Isupov, M.N., Littlechild, J.A., Sun, L., Liu, X., Wang, Q., Gong, H., Dong, P., Zhang, N., Wu, Y.(2021) J Agric Food Chem 69: 1079-1090
- PubMed: 33445864
- DOI: https://doi.org/10.1021/acs.jafc.0c06338
- Primary Citation of Related Structures:
7C4D - PubMed Abstract:
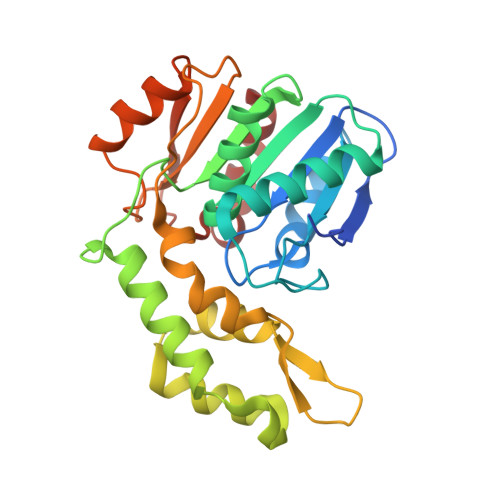
Lipolytic enzymes are essential biocatalysts in food processing as well as pharmaceutical and pesticide industries, catalyzing the cleavage of ester bonds in a variety of acyl chain substrates. Here, we report the crystal structure of an esterase from the deep-sea hydrothermal vent of the East Pacific Rise (EprEst). The X-ray structure of EprEst in complex with the ligand, acetate, has been determined at 2.03 Å resolution. The structure reveals a unique spatial arrangement and orientation of the helix cap domain and α/β hydrolase domain, which form a substrate pocket with preference for short-chain acyl groups. Molecular docking analysis further demonstrated that the active site pocket could accommodate p -nitrophenyl ( p NP) carboxyl ligands of varying lengths (≤6 C atoms), with p NP-butyrate ester predicted to have the highest binding affinity. Additionally, the semirational design was conducted to improve the thermostability of EprEst by enzyme engineering based on the established structure and multiple sequence alignment. A mutation, K114P, introduced in the hinge region of the esterase, which displayed increased thermostability and enzyme activity. Collectively, the structural and functional data obtained herein could be used as basis for further protein engineering to ultimately expand the scope of industrial applications of marine-derived lipolytic enzymes.
- State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fuzhou 350002, China.
Organizational Affiliation: