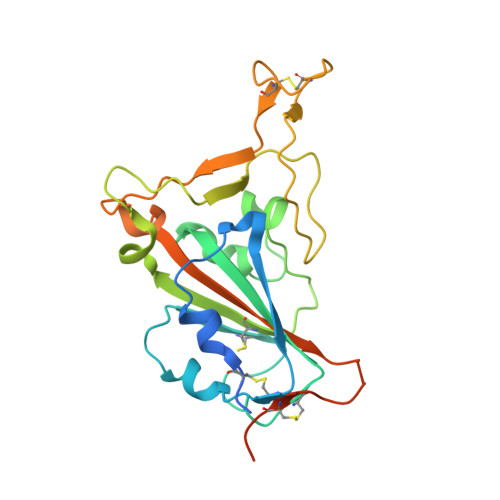
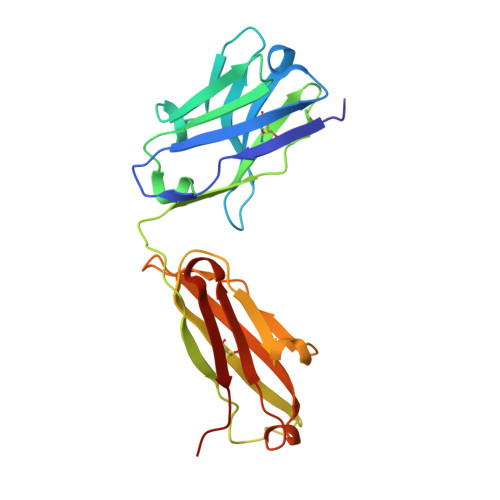
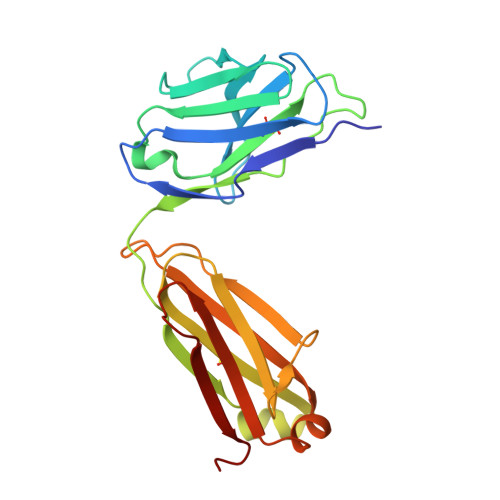
A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Wu, Y., Wang, F., Shen, C., Peng, W., Li, D., Zhao, C., Li, Z., Li, S., Bi, Y., Yang, Y., Gong, Y., Xiao, H., Fan, Z., Tan, S., Wu, G., Tan, W., Lu, X., Fan, C., Wang, Q., Liu, Y., Zhang, C., Qi, J., Gao, G.F., Gao, F., Liu, L.(2020) Science 368: 1274-1278
- PubMed: 32404477
- DOI: https://doi.org/10.1126/science.abc2241
- Primary Citation of Related Structures:
7BZ5 - PubMed Abstract:
Neutralizing antibodies could potentially be used as antivirals against the coronavirus disease 2019 (COVID-19) pandemic. Here, we report isolation of four human-origin monoclonal antibodies from a convalescent patient, all of which display neutralization abilities. The antibodies B38 and H4 block binding between the spike glycoprotein receptor binding domain (RBD) of the virus and the cellular receptor angiotensin-converting enzyme 2 (ACE2). A competition assay indicated different epitopes on the RBD for these two antibodies, making them a potentially promising virus-targeting monoclonal antibody pair for avoiding immune escape in future clinical applications. Moreover, a therapeutic study in a mouse model validated that these antibodies can reduce virus titers in infected lungs. The RBD-B38 complex structure revealed that most residues on the epitope overlap with the RBD-ACE2 binding interface, explaining the blocking effect and neutralizing capacity. Our results highlight the promise of antibody-based therapeutics and provide a structural basis for rational vaccine design.
- Department of Pathogen Microbiology, School of Basic Medical Sciences, Capital Medical University, Beijing, China. wuy@biols.ac.cn gaofeng@tib.cas.cn gaof@im.ac.cn liulei3322@aliyun.com.
Organizational Affiliation: