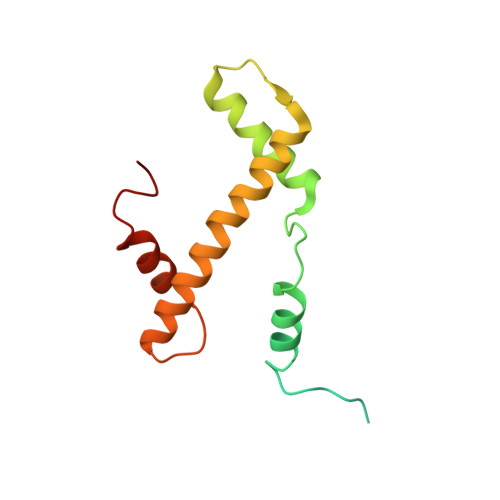
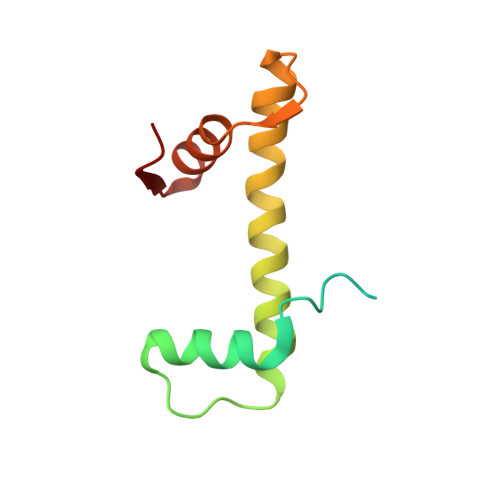
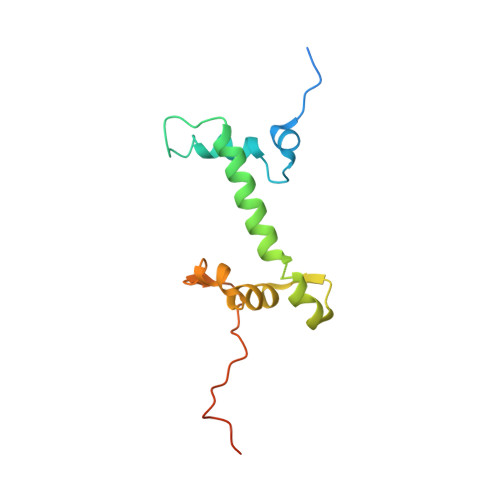
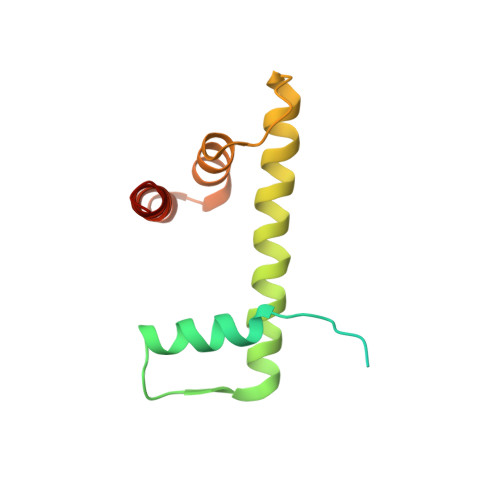
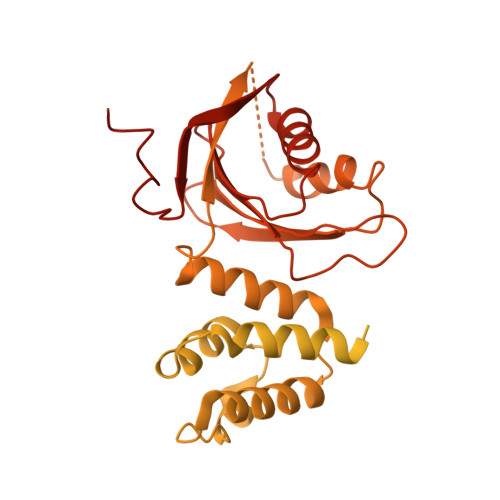
Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Ariyoshi, M., Makino, F., Watanabe, R., Nakagawa, R., Kato, T., Namba, K., Arimura, Y., Fujita, R., Kurumizaka, H., Okumura, E.I., Hara, M., Fukagawa, T.(2021) EMBO J 40: e105671-e105671
- PubMed: 33463726
- DOI: https://doi.org/10.15252/embj.2020105671
- Primary Citation of Related Structures:
7BXT, 7BY0 - PubMed Abstract:
The CENP-A nucleosome is a key structure for kinetochore assembly. Once the CENP-A nucleosome is established in the centromere, additional proteins recognize the CENP-A nucleosome to form a kinetochore. CENP-C and CENP-N are CENP-A binding proteins. We previously demonstrated that vertebrate CENP-C binding to the CENP-A nucleosome is regulated by CDK1-mediated CENP-C phosphorylation. However, it is still unknown how the phosphorylation of CENP-C regulates its binding to CENP-A. It is also not completely understood how and whether CENP-C and CENP-N act together on the CENP-A nucleosome. Here, using cryo-electron microscopy (cryo-EM) in combination with biochemical approaches, we reveal a stable CENP-A nucleosome-binding mode of CENP-C through unique regions. The chicken CENP-C structure bound to the CENP-A nucleosome is stabilized by an intramolecular link through the phosphorylated CENP-C residue. The stable CENP-A-CENP-C complex excludes CENP-N from the CENP-A nucleosome. These findings provide mechanistic insights into the dynamic kinetochore assembly regulated by CDK1-mediated CENP-C phosphorylation.
- Graduate School of Frontier Biosciences, Osaka University, Suita, Osaka, Japan.
Organizational Affiliation: