Crystal structure of MurE from E.coli
Koekemoer, L., Steindel, M., Fairhead, M., Talon, R., Douangamath, A., Arrowsmith, C.H., Edwards, A.M., Bountra, C., von Delft, F., Krojer, T.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | A [auth AAA], B [auth BBB] | 496 | Escherichia coli | Mutation(s): 0 Gene Names: murE, GHR40_01000, NCTC12650_04426 EC: 6.3.2.13 | 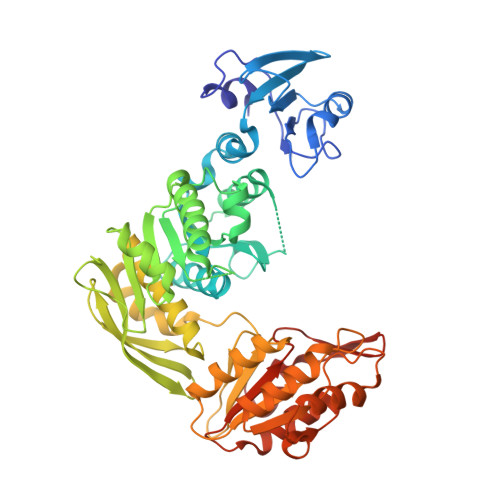 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NUJ (Subject of Investigation/LOI) Query on NUJ | D [auth BBB] | trans-3-[(2,6-dimethylpyrimidin-4-yl)(methyl)amino]cyclobutan-1-ol C11 H17 N3 O SEZOHQAJJCEOMQ-MGCOHNPYSA-N |  | ||
| DMS Query on DMS | C [auth AAA], E [auth BBB] | DIMETHYL SULFOXIDE C2 H6 O S IAZDPXIOMUYVGZ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 58.276 | α = 97.234 |
| b = 58.293 | β = 91.447 |
| c = 74.115 | γ = 105.313 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| xia2 | data reduction |
| xia2 | data scaling |
| PHASER | phasing |