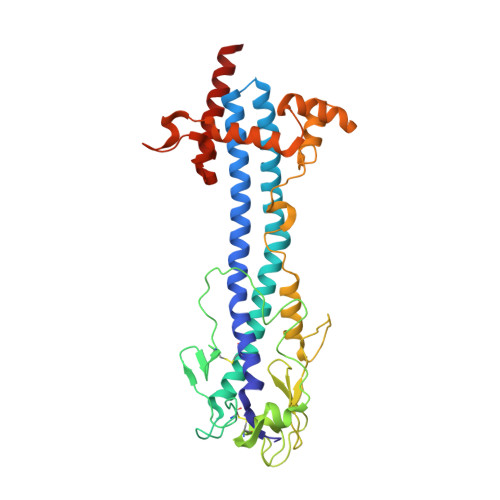
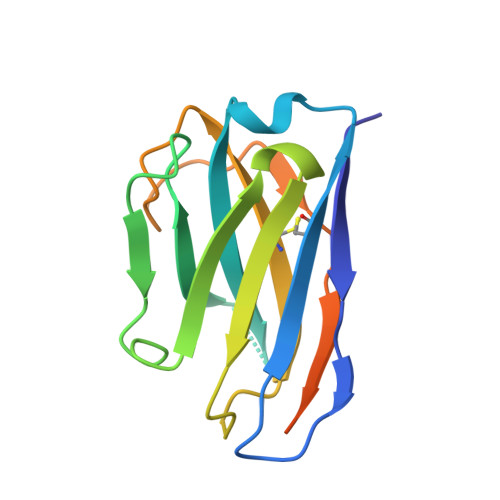
Nanobody-mediated macromolecular crowding induces membrane fission and remodeling in the African trypanosome.
Hempelmann, A., Hartleb, L., van Straaten, M., Hashemi, H., Zeelen, J.P., Bongers, K., Papavasiliou, F.N., Engstler, M., Stebbins, C.E., Jones, N.G.(2021) Cell Rep 37: 109923-109923
- PubMed: 34731611
- DOI: https://doi.org/10.1016/j.celrep.2021.109923
- Primary Citation of Related Structures:
7AQX, 7AQY, 7AQZ, 7AR0 - PubMed Abstract:
The dense variant surface glycoprotein (VSG) coat of African trypanosomes represents the primary host-pathogen interface. Antigenic variation prevents clearing of the pathogen by employing a large repertoire of antigenically distinct VSG genes, thus neutralizing the host's antibody response. To explore the epitope space of VSGs, we generate anti-VSG nanobodies and combine high-resolution structural analysis of VSG-nanobody complexes with binding assays on living cells, revealing that these camelid antibodies bind deeply inside the coat. One nanobody causes rapid loss of cellular motility, possibly due to blockage of VSG mobility on the coat, whose rapid endocytosis and exocytosis are mechanistically linked to Trypanosoma brucei propulsion and whose density is required for survival. Electron microscopy studies demonstrate that this loss of motility is accompanied by rapid formation and shedding of nanovesicles and nanotubes, suggesting that increased protein crowding on the dense membrane can be a driving force for membrane fission in living cells.
- Division of Structural Biology of Infection and Immunity, German Cancer Research Center, Heidelberg 69120, Germany.
Organizational Affiliation: