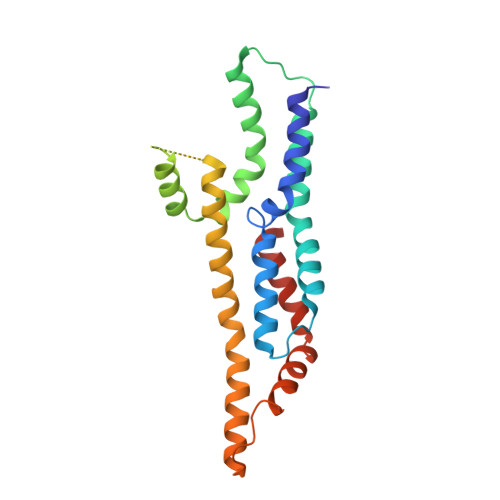
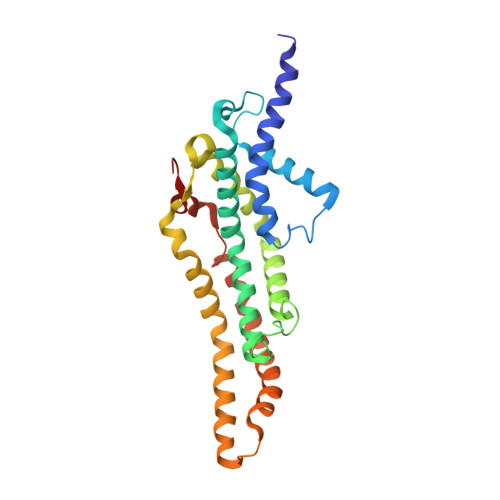
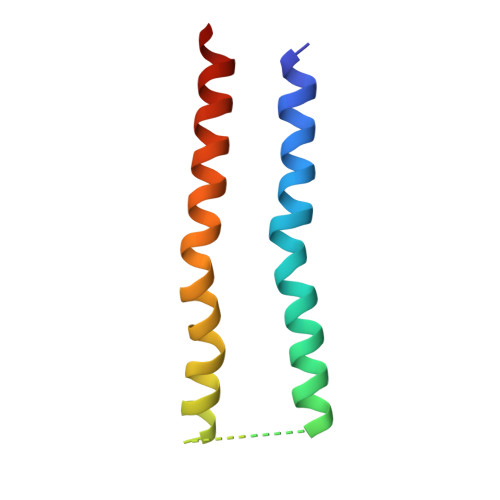
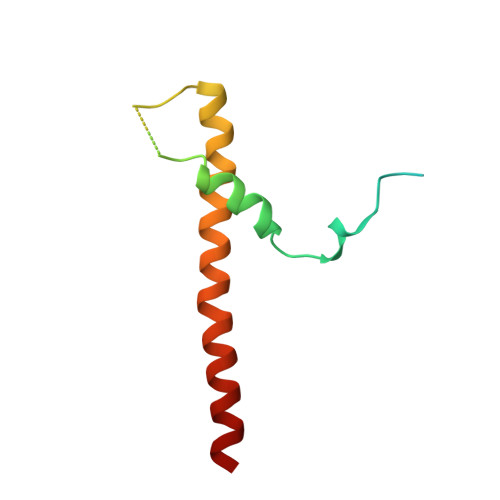
Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation.
Miletic, S., Fahrenkamp, D., Goessweiner-Mohr, N., Wald, J., Pantel, M., Vesper, O., Kotov, V., Marlovits, T.C.(2021) Nat Commun 12: 1546-1546
- PubMed: 33750771
- DOI: https://doi.org/10.1038/s41467-021-21143-1
- Primary Citation of Related Structures:
7AGX - PubMed Abstract:
Many bacterial pathogens rely on virulent type III secretion systems (T3SSs) or injectisomes to translocate effector proteins in order to establish infection. The central component of the injectisome is the needle complex which assembles a continuous conduit crossing the bacterial envelope and the host cell membrane to mediate effector protein translocation. However, the molecular principles underlying type III secretion remain elusive. Here, we report a structure of an active Salmonella enterica serovar Typhimurium needle complex engaged with the effector protein SptP in two functional states, revealing the complete 800Å-long secretion conduit and unraveling the critical role of the export apparatus (EA) subcomplex in type III secretion. Unfolded substrates enter the EA through a hydrophilic constriction formed by SpaQ proteins, which enables side chain-independent substrate transport. Above, a methionine gasket formed by SpaP proteins functions as a gate that dilates to accommodate substrates while preventing leaky pore formation. Following gate penetration, a moveable SpaR loop first folds up to then support substrate transport. Together, these findings establish the molecular basis for substrate translocation through T3SSs and improve our understanding of bacterial pathogenicity and motility.
- University Medical Center Hamburg-Eppendorf (UKE), Institute of Structural and Systems Biology, Hamburg, Germany.
Organizational Affiliation: