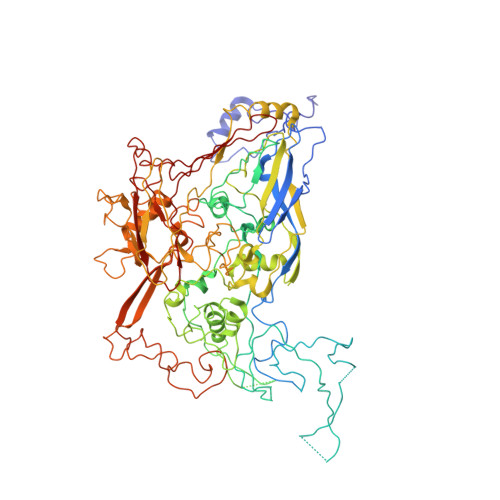
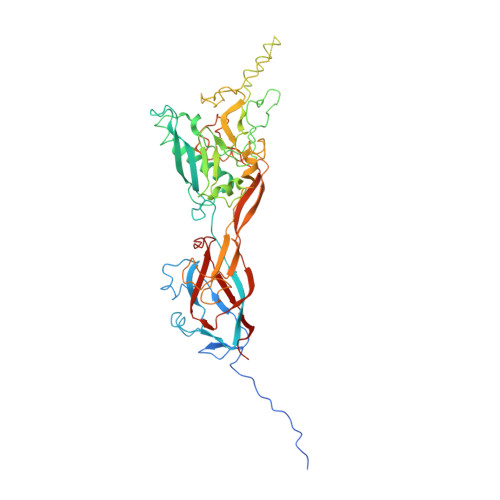
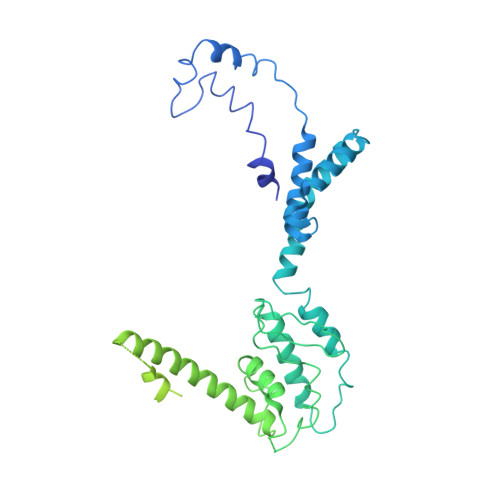
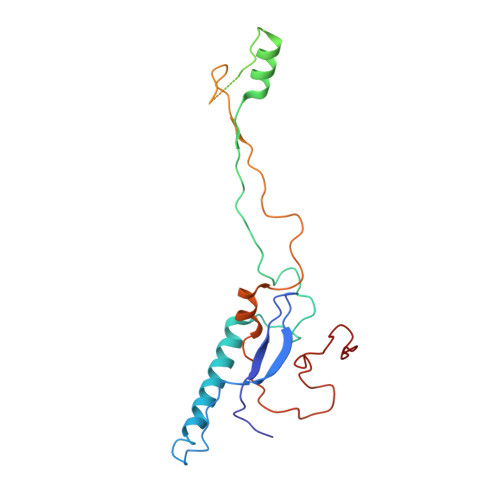
The structure of enteric human adenovirus 41-A leading cause of diarrhea in children.
Rafie, K., Lenman, A., Fuchs, J., Rajan, A., Arnberg, N., Carlson, L.A.(2021) Sci Adv 7
- PubMed: 33523995
- DOI: https://doi.org/10.1126/sciadv.abe0974
- Primary Citation of Related Structures:
6Z7N, 6Z7Q - PubMed Abstract:
Human adenovirus (HAdV) types F40 and F41 are a prominent cause of diarrhea and diarrhea-associated mortality in young children worldwide. These enteric HAdVs differ notably in tissue tropism and pathogenicity from respiratory and ocular adenoviruses, but the structural basis for this divergence has been unknown. Here, we present the first structure of an enteric HAdV-HAdV-F41-determined by cryo-electron microscopy to a resolution of 3.8 Å. The structure reveals extensive alterations to the virion exterior as compared to nonenteric HAdVs, including a unique arrangement of capsid protein IX. The structure also provides new insights into conserved aspects of HAdV architecture such as a proposed location of core protein V, which links the viral DNA to the capsid, and assembly-induced conformational changes in the penton base protein. Our findings provide the structural basis for adaptation of enteric HAdVs to a fundamentally different tissue tropism.
- Department of Medical Biochemistry and Biophysics, Umeå University, Umeå, Sweden.
Organizational Affiliation: