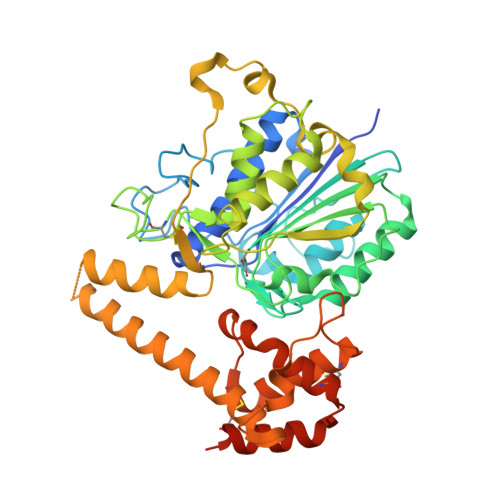
Structural and functional studies ofArabidopsis thalianalegumain beta reveal isoform specific mechanisms of activation and substrate recognition.
Dall, E., Zauner, F.B., Soh, W.T., Demir, F., Dahms, S.O., Cabrele, C., Huesgen, P.F., Brandstetter, H.(2020) J Biological Chem 295: 13047-13064
- PubMed: 32719006
- DOI: https://doi.org/10.1074/jbc.RA120.014478
- Primary Citation of Related Structures:
6YSA - PubMed Abstract:
The vacuolar cysteine protease legumain plays important functions in seed maturation and plant programmed cell death. Because of their dual protease and ligase activity, plant legumains have become of particular biotechnological interest, e.g. for the synthesis of cyclic peptides for drug design or for protein engineering. However, the molecular mechanisms behind their dual protease and ligase activities are still poorly understood, limiting their applications. Here, we present the crystal structure of Arabidopsis thaliana legumain isoform β (AtLEGβ) in its zymogen state. Combining structural and biochemical experiments, we show for the first time that plant legumains encode distinct, isoform-specific activation mechanisms. Whereas the autocatalytic activation of isoform γ (AtLEGγ) is controlled by the latency-conferring dimer state, the activation of the monomeric AtLEGβ is concentration independent. Additionally, in AtLEGβ the plant-characteristic two-chain intermediate state is stabilized by hydrophobic rather than ionic interactions, as in AtLEGγ, resulting in significantly different pH stability profiles. The crystal structure of AtLEGβ revealed unrestricted nonprime substrate binding pockets, consistent with the broad substrate specificity, as determined by degradomic assays. Further to its protease activity, we show that AtLEGβ exhibits a true peptide ligase activity. Whereas cleavage-dependent transpeptidase activity has been reported for other plant legumains, AtLEGβ is the first example of a plant legumain capable of linking free termini. The discovery of these isoform-specific differences will allow us to identify and rationally design efficient ligases with application in biotechnology and drug development.
- Department of Biosciences, University of Salzburg, Salzburg, Austria. Electronic address: elfriede.dall@sbg.ac.at.
Organizational Affiliation: