FGCaMP7, an Improved Version of Fungi-Based Ratiometric Calcium Indicator for In Vivo Visualization of Neuronal Activity.
Barykina, N.V., Sotskov, V.P., Gruzdeva, A.M., Wu, Y.K., Portugues, R., Subach, O.M., Chefanova, E.S., Plusnin, V.V., Ivashkina, O.I., Anokhin, K.V., Vlaskina, A.V., Korzhenevskiy, D.A., Nikolaeva, A.Y., Boyko, K.M., Rakitina, T.V., Varizhuk, A.M., Pozmogova, G.E., Subach, F.V.(2020) Int J Mol Sci 21
- PubMed: 32344594
- DOI: https://doi.org/10.3390/ijms21083012
- Primary Citation of Related Structures:
6XU4 - PubMed Abstract:
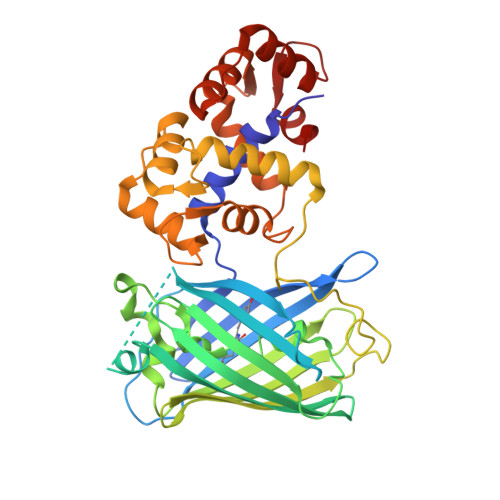
Genetically encoded calcium indicators (GECIs) have become a widespread tool for the visualization of neuronal activity. As compared to popular GCaMP GECIs, the FGCaMP indicator benefits from calmodulin and M13-peptide from the fungi Aspergillus niger and Aspergillus fumigatus , which prevent its interaction with the intracellular environment. However, FGCaMP exhibits a two-phase fluorescence behavior with the variation of calcium ion concentration, has moderate sensitivity in neurons (as compared to the GCaMP6s indicator), and has not been fully characterized in vitro and in vivo. To address these limitations, we developed an enhanced version of FGCaMP, called FGCaMP7. FGCaMP7 preserves the ratiometric phenotype of FGCaMP, with a 3.1-fold larger ratiometric dynamic range in vitro. FGCaMP7 demonstrates 2.7- and 8.7-fold greater photostability compared to mEGFP and mTagBFP2 fluorescent proteins in vitro, respectively. The ratiometric response of FGCaMP7 is 1.6- and 1.4-fold higher, compared to the intensiometric response of GCaMP6s, in non-stimulated and stimulated neuronal cultures, respectively. We reveal the inertness of FGCaMP7 to the intracellular environment of HeLa cells using its truncated version with a deleted M13-like peptide; in contrast to the similarly truncated variant of GCaMP6s. We characterize the crystal structure of the parental FGCaMP indicator. Finally, we test the in vivo performance of FGCaMP7 in mouse brain using a two-photon microscope and an NVista miniscope; and in zebrafish using two-color ratiometric confocal imaging.
- Laboratory for Neurobiology of Memory, P.K. Anokhin Research Institute of Normal Physiology, 125315 Moscow, Russia.
Organizational Affiliation: