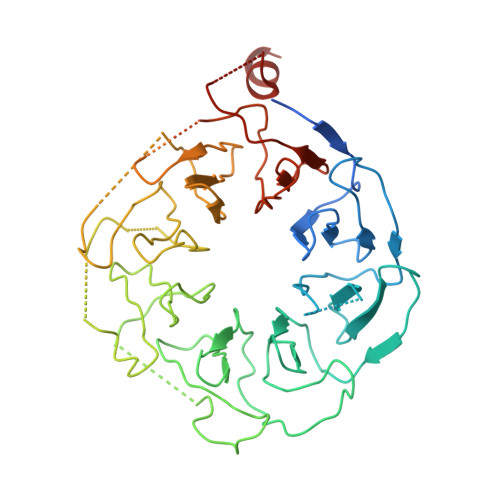
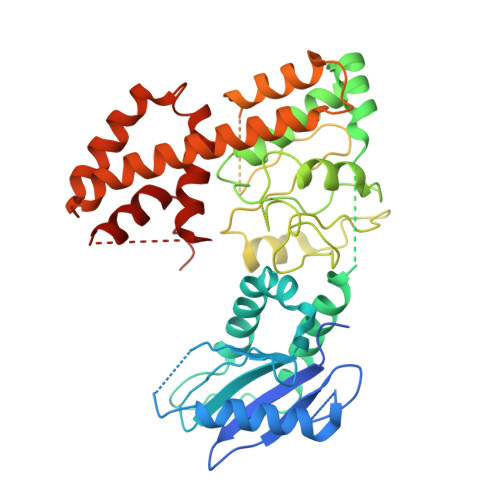
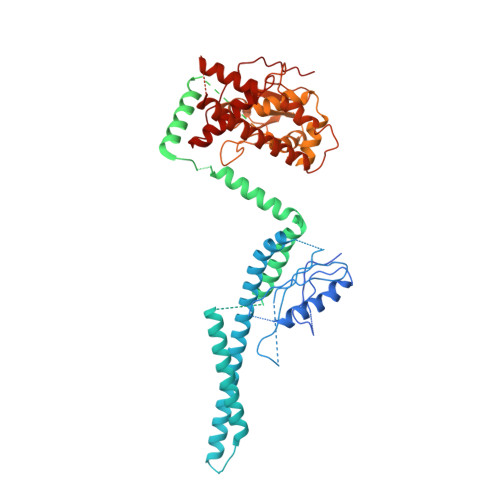
Structure of the C9orf72 ARF GAP complex that is haploinsufficient in ALS and FTD.
Su, M.Y., Fromm, S.A., Zoncu, R., Hurley, J.H.(2020) Nature 585: 251-255
- PubMed: 32848248
- DOI: https://doi.org/10.1038/s41586-020-2633-x
- Primary Citation of Related Structures:
6V4U - PubMed Abstract:
Mutation of C9orf72 is the most prevalent defect associated with amyotrophic lateral sclerosis and frontotemporal degeneration 1 . Together with hexanucleotide-repeat expansion 2,3 , haploinsufficiency of C9orf72 contributes to neuronal dysfunction 4-6 . Here we determine the structure of the C9orf72-SMCR8-WDR41 complex by cryo-electron microscopy. C9orf72 and SMCR8 both contain longin and DENN (differentially expressed in normal and neoplastic cells) domains 7 , and WDR41 is a β-propeller protein that binds to SMCR8 such that the whole structure resembles an eye slip hook. Contacts between WDR41 and the DENN domain of SMCR8 drive the lysosomal localization of the complex in conditions of amino acid starvation. The structure suggested that C9orf72-SMCR8 is a GTPase-activating protein (GAP), and we found that C9orf72-SMCR8-WDR41 acts as a GAP for the ARF family of small GTPases. These data shed light on the function of C9orf72 in normal physiology, and in amyotrophic lateral sclerosis and frontotemporal degeneration.
- Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA, USA.
Organizational Affiliation: