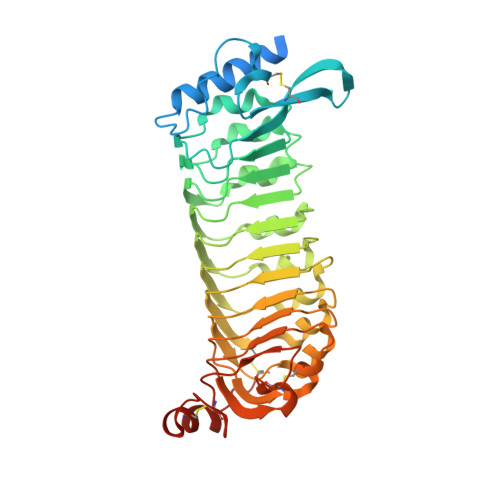
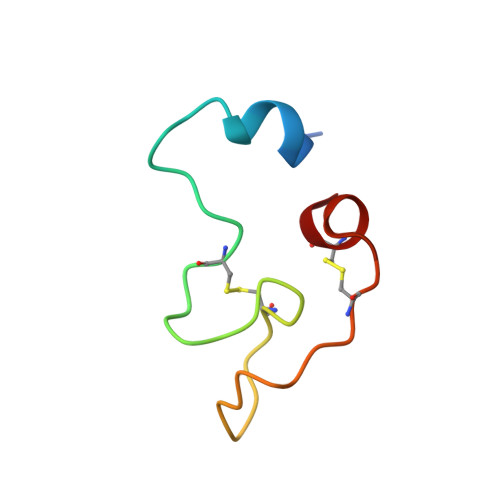
Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth.
Moussu, S., Broyart, C., Santos-Fernandez, G., Augustin, S., Wehrle, S., Grossniklaus, U., Santiago, J.(2020) Proc Natl Acad Sci U S A 117: 7494-7503
- PubMed: 32165538
- DOI: https://doi.org/10.1073/pnas.2000100117
- Primary Citation of Related Structures:
6QWN, 6QXP, 6TME - PubMed Abstract:
Plant reproduction relies on the highly regulated growth of the pollen tube for sperm delivery. This process is controlled by secreted RALF signaling peptides, which have previously been shown to be perceived by Catharanthus roseus RLK1-like ( Cr RLK1Ls) membrane receptor-kinases/LORELEI-like GLYCOLPHOSPHATIDYLINOSITOL (GPI)-ANCHORED PROTEINS (LLG) complexes, or by leucine-rich repeat (LRR) extensin proteins (LRXs). Here, we demonstrate that RALF peptides fold into bioactive, disulfide bond-stabilized proteins that bind the LRR domain of LRX proteins with low nanomolar affinity. Crystal structures of LRX2-RALF4 and LRX8-RALF4 complexes at 3.2- and 3.9-Å resolution, respectively, reveal a dimeric arrangement of LRX proteins, with each monomer binding one folded RALF peptide. Structure-based mutations targeting the LRX-RALF4 complex interface, or the RALF4 fold, reduce RALF4 binding to LRX8 in vitro and RALF4 function in growing pollen tubes. Mutants targeting the disulfide-bond stabilized LRX dimer interface fail to rescue lrx infertility phenotypes. Quantitative biochemical assays reveal that RALF4 binds LLGs and LRX cell-wall modules with drastically different binding affinities, and with distinct and mutually exclusive binding modes. Our biochemical, structural, and genetic analyses reveal a complex signaling network by which RALF ligands instruct different signaling proteins using distinct targeting mechanisms.
- The Plant Signaling Mechanisms Laboratory, Department of Plant Molecular Biology, University of Lausanne, 1015 Lausanne, Switzerland.
Organizational Affiliation: