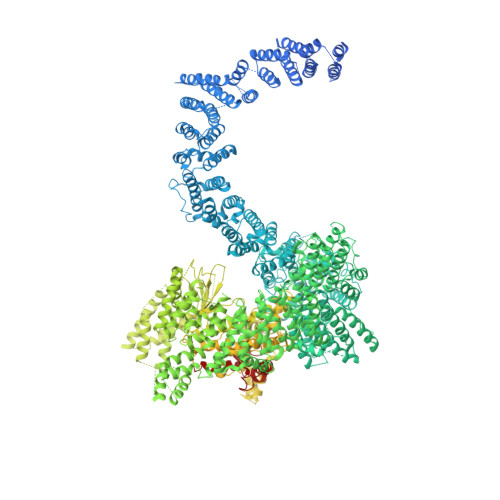
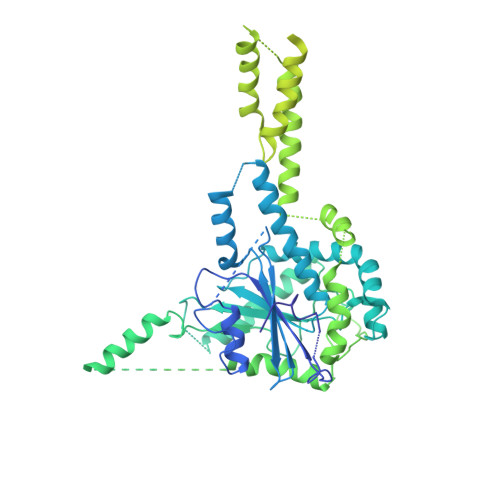
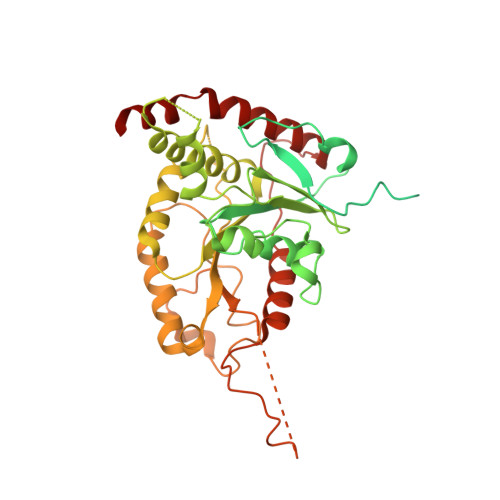
InsP6binding to PIKK kinases revealed by the cryo-EM structure of an SMG1-SMG8-SMG9 complex.
Gat, Y., Schuller, J.M., Lingaraju, M., Weyher, E., Bonneau, F., Strauss, M., Murray, P.J., Conti, E.(2019) Nat Struct Mol Biol 26: 1089-1093
- PubMed: 31792449
- DOI: https://doi.org/10.1038/s41594-019-0342-7
- Primary Citation of Related Structures:
6SYT - PubMed Abstract:
We report the 3.45-Å resolution cryo-EM structure of human SMG1-SMG8-SMG9, a phosphatidylinositol-3-kinase (PI(3)K)-related protein kinase (PIKK) complex central to messenger RNA surveillance. Structural and MS analyses reveal the presence of inositol hexaphosphate (InsP 6 ) in the SMG1 kinase. We show that the InsP 6 -binding site is conserved in mammalian target of rapamycin (mTOR) and potentially other PIKK members, and that it is required for optimal in vitro phosphorylation of both SMG1 and mTOR substrates.
- Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Munich, Germany.
Organizational Affiliation: