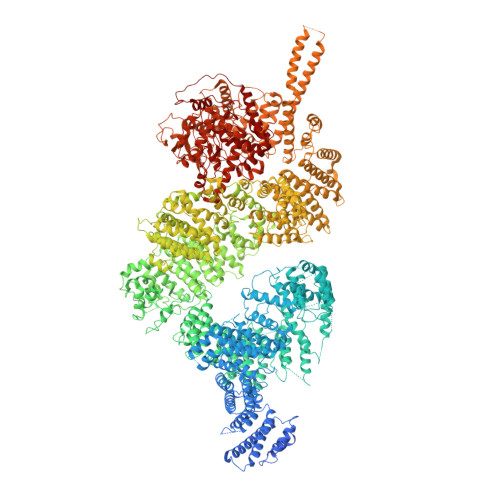
Cryo-EM Structure of Nucleotide-Bound Tel1ATMUnravels the Molecular Basis of Inhibition and Structural Rationale for Disease-Associated Mutations.
Yates, L.A., Williams, R.M., Hailemariam, S., Ayala, R., Burgers, P., Zhang, X.(2020) Structure 28: 96-104.e3
- PubMed: 31740029
- DOI: https://doi.org/10.1016/j.str.2019.10.012
- Primary Citation of Related Structures:
6S8F - PubMed Abstract:
Yeast Tel1 and its highly conserved human ortholog ataxia-telangiectasia mutated (ATM) are large protein kinases central to the maintenance of genome integrity. Mutations in ATM are found in ataxia-telangiectasia (A-T) patients and ATM is one of the most frequently mutated genes in many cancers. Using cryoelectron microscopy, we present the structure of Tel1 in a nucleotide-bound state. Our structure reveals molecular details of key residues surrounding the nucleotide binding site and provides a structural and molecular basis for its intrinsically low basal activity. We show that the catalytic residues are in a productive conformation for catalysis, but the phosphatidylinositol 3-kinase-related kinase (PIKK) regulatory domain insert restricts peptide substrate access and the N-lobe is in an open conformation, thus explaining the requirement for Tel1 activation. Structural comparisons with other PIKKs suggest a conserved and common allosteric activation mechanism. Our work also provides a structural rationale for many mutations found in A-T and cancer.
- Section of Structural and Synthetic Biology, Faculty of Infectious Diseases, Imperial College London, London SW7 2AZ, UK.
Organizational Affiliation: