Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Bhat, V.D., McCann, K.L., Wang, Y., Fonseca, D.R., Shukla, T., Alexander, J.C., Qiu, C., Wickens, M., Lo, T.W., Tanaka Hall, T.M., Campbell, Z.T.(2019) Elife 8
- PubMed: 30652968
- DOI: https://doi.org/10.7554/eLife.43788
- Primary Citation of Related Structures:
6NOC, 6NOD, 6NOF, 6NOH - PubMed Abstract:
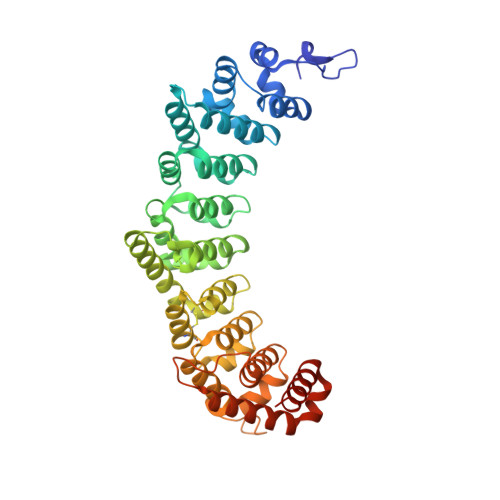
PUF ( PU milio/ F BF) RNA-binding proteins recognize distinct elements. In C. elegans , PUF-8 binds to an 8-nt motif and restricts proliferation in the germline. Conversely, FBF-2 recognizes a 9-nt element and promotes mitosis. To understand how motif divergence relates to biological function, we first determined a crystal structure of PUF-8. Comparison of this structure to that of FBF-2 revealed a major difference in a central repeat. We devised a modified yeast 3-hybrid screen to identify mutations that confer recognition of an 8-nt element to FBF-2. We identified several such mutants and validated structurally and biochemically their binding to 8-nt RNA elements. Using genome engineering, we generated a mutant animal with a substitution in FBF-2 that confers preferential binding to the PUF-8 element. The mutant largely rescued overproliferation in animals that spontaneously generate tumors in the absence of puf-8 . This work highlights the critical role of motif length in the specification of biological function.
- Department of Biological Sciences, University of Texas Dallas, Richardson, United States.
Organizational Affiliation: