Discovery of the Universal tRNA Binding Mode for the TsaD-like Components of the t6A tRNA Modification Pathway
Stec, B.(2023) Biophysica 3: 288-306
Experimental Data Snapshot
Starting Model: experimental
View more details
This entry reflects an alternative modeling of the original data in: 6FPE
(2023) Biophysica 3: 288-306
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| tRNA threonylcarbamoyladenosine biosynthesis protein TsaB | A, D [auth C] | 206 | Thermotoga maritima MSB8 | Mutation(s): 0 Gene Names: tsaB, TM_0874, Tmari_0876 | 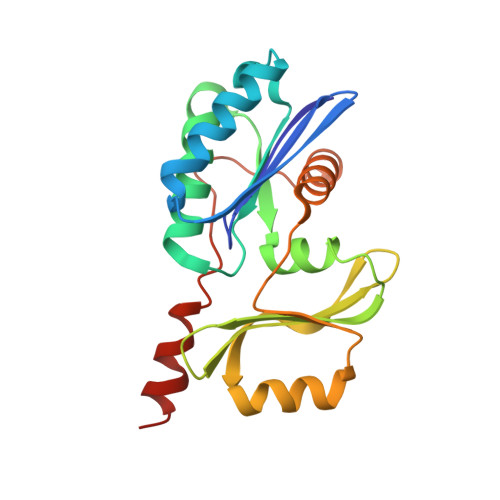 |
UniProt | |||||
Find proteins for Q9WZX7 (Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8)) Explore Q9WZX7 Go to UniProtKB: Q9WZX7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9WZX7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| tRNA N6-adenosine threonylcarbamoyltransferase | B, E [auth G] | 327 | Thermotoga maritima MSB8 | Mutation(s): 0 Gene Names: tsaD, gcp, TM_0145 EC: 2.3.1.234 | 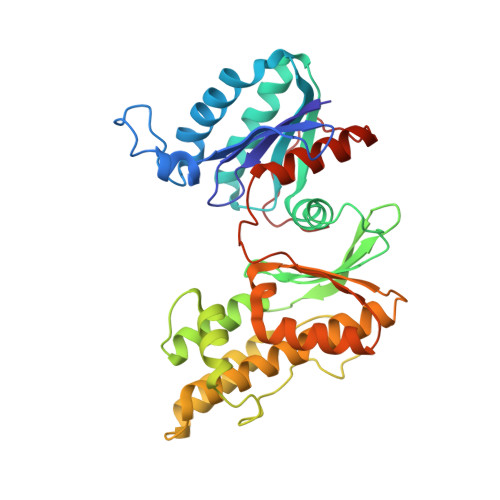 |
UniProt | |||||
Find proteins for Q9WXZ2 (Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8)) Explore Q9WXZ2 Go to UniProtKB: Q9WXZ2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9WXZ2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TsaE | C [auth D], F [auth E] | 184 | Thermotoga maritima MSB8 | Mutation(s): 0 Gene Names: TsaE | 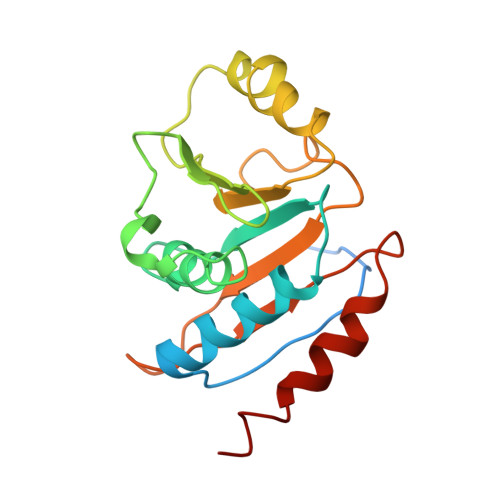 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| APC Query on APC | J [auth D], N [auth E] | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER C11 H18 N5 O12 P3 CAWZRIXWFRFUQB-IOSLPCCCSA-N |  | ||
| TXA Query on TXA | G [auth B], K [auth G] | threonylcarbamoyladenylate C15 H21 N6 O11 P GHLUPQUHEIJRCU-DWVDDHQFSA-N |  | ||
| ZN Query on ZN | H [auth B], L [auth G] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | I [auth D], M [auth E] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 84.31 | α = 90 |
| b = 113.99 | β = 90 |
| c = 177.57 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |