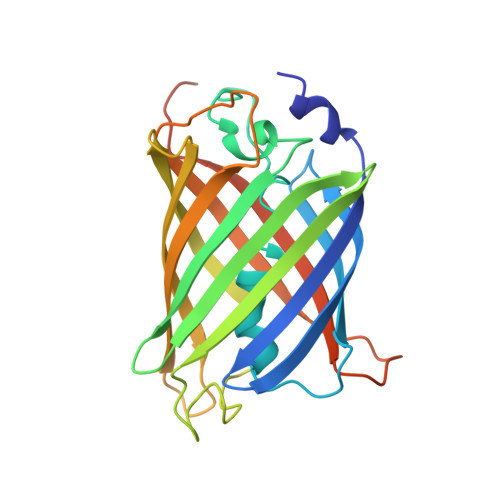
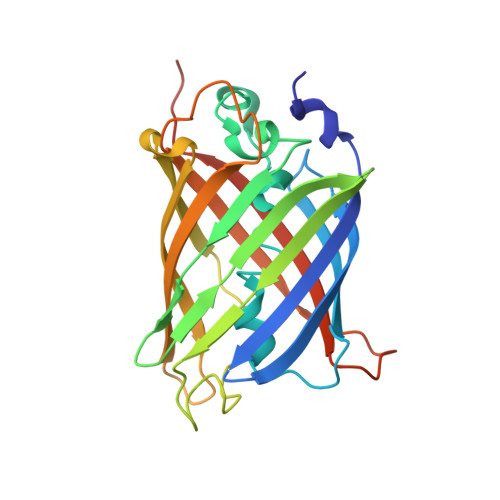
Supercharging enables organized assembly of synthetic biomolecules.
Simon, A.J., Zhou, Y., Ramasubramani, V., Glaser, J., Pothukuchy, A., Gollihar, J., Gerberich, J.C., Leggere, J.C., Morrow, B.R., Jung, C., Glotzer, S.C., Taylor, D.W., Ellington, A.D.(2019) Nat Chem 11: 204-212
- PubMed: 30643229
- DOI: https://doi.org/10.1038/s41557-018-0196-3
- Primary Citation of Related Structures:
6MDR - PubMed Abstract:
Symmetrical protein oligomers are ubiquitous in biological systems and perform key structural and regulatory functions. However, there are few methods for constructing such oligomers. Here we have engineered completely synthetic, symmetrical oligomers by combining pairs of oppositely supercharged variants of a normally monomeric model protein through a strategy we term 'supercharged protein assembly' (SuPrA). We show that supercharged variants of green fluorescent protein can assemble into a variety of architectures including a well-defined symmetrical 16-mer structure that we solved using cryo-electron microscopy at 3.47 Å resolution. The 16-mer is composed of two stacked rings of octamers, in which the octamers contain supercharged proteins of alternating charges, and interactions within and between the rings are mediated by a variety of specific electrostatic contacts. The ready assembly of this structure suggests that combining oppositely supercharged pairs of protein variants may provide broad opportunities for generating novel architectures via otherwise unprogrammed interactions.
- Center for Systems and Synthetic Biology, University of Texas at Austin, Austin, TX, USA.
Organizational Affiliation: