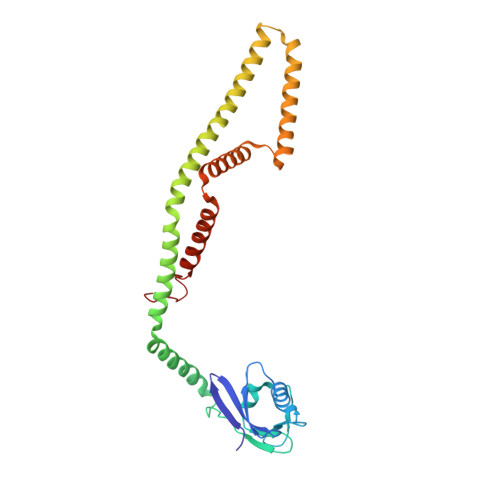
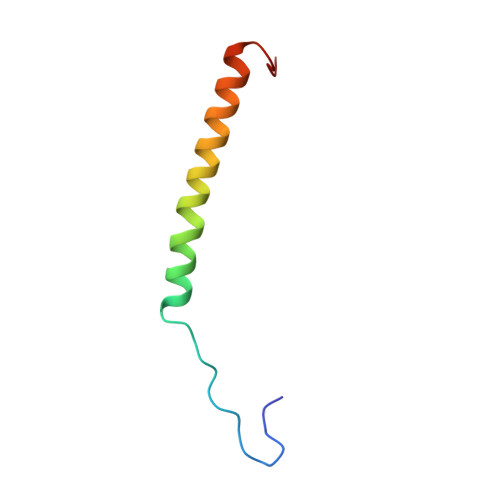
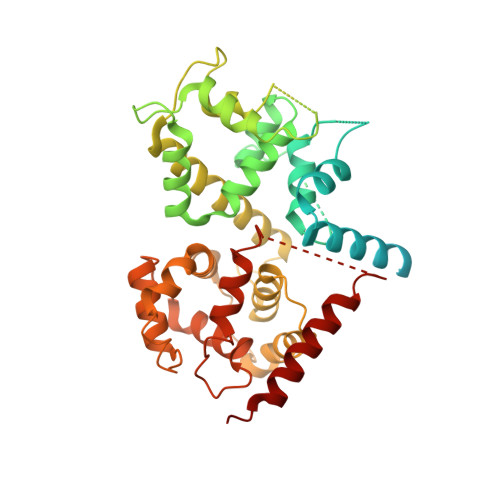
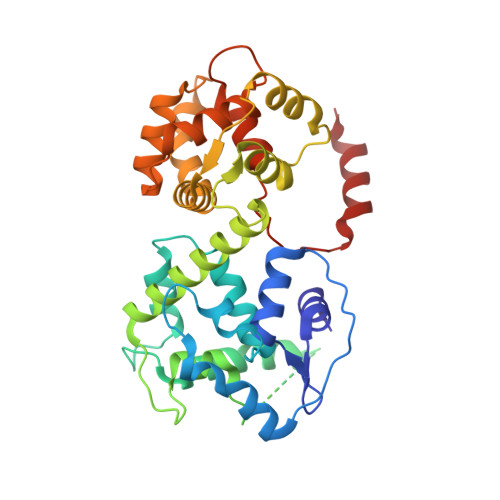
Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Zhuo, W., Zhou, H., Guo, R., Yi, J., Zhang, L., Yu, L., Sui, Y., Zeng, W., Wang, P., Yang, M.(2021) Protein Cell 12: 220-229
- PubMed: 32862359
- DOI: https://doi.org/10.1007/s13238-020-00776-w
- Primary Citation of Related Structures:
6K7X, 6K7Y - Ministry of Education Key Laboratory of Protein Science, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing, 100084, China. zhuowei@tsinghua.edu.cn.
Organizational Affiliation: