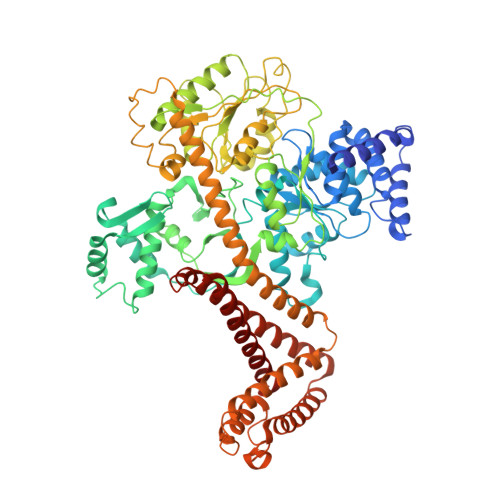
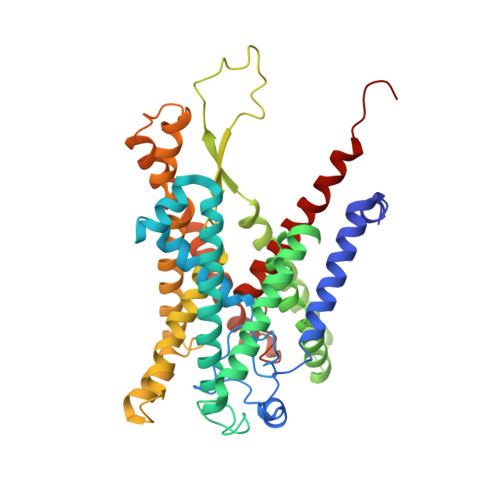
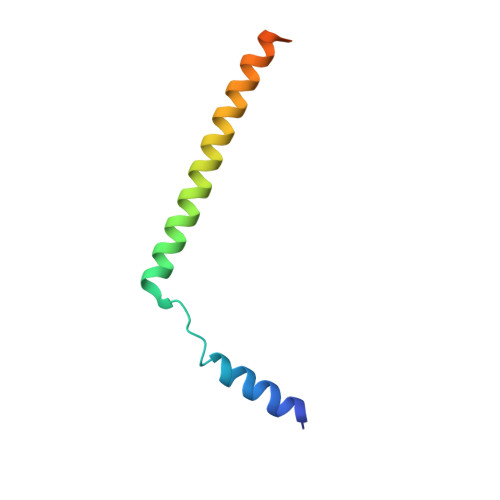
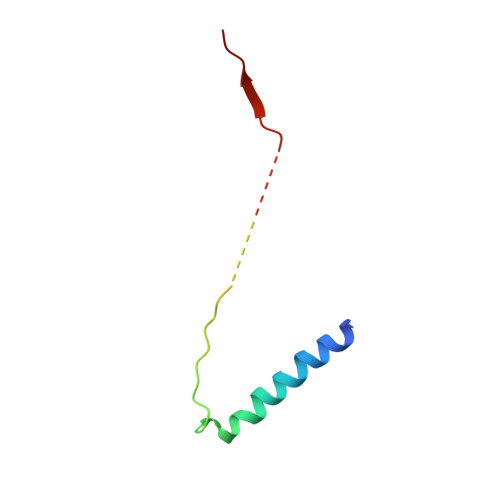
Structure of the substrate-engaged SecA-SecY protein translocation machine.
Ma, C., Wu, X., Sun, D., Park, E., Catipovic, M.A., Rapoport, T.A., Gao, N., Li, L.(2019) Nat Commun 10: 2872-2872
- PubMed: 31253804
- DOI: https://doi.org/10.1038/s41467-019-10918-2
- Primary Citation of Related Structures:
6ITC - PubMed Abstract:
The Sec61/SecY channel allows the translocation of many proteins across the eukaryotic endoplasmic reticulum membrane or the prokaryotic plasma membrane. In bacteria, most secretory proteins are transported post-translationally through the SecY channel by the SecA ATPase. How a polypeptide is moved through the SecA-SecY complex is poorly understood, as structural information is lacking. Here, we report an electron cryo-microscopy (cryo-EM) structure of a translocating SecA-SecY complex in a lipid environment. The translocating polypeptide chain can be traced through both SecA and SecY. In the captured transition state of ATP hydrolysis, SecA's two-helix finger is close to the polypeptide, while SecA's clamp interacts with the polypeptide in a sequence-independent manner by inducing a short β-strand. Taking into account previous biochemical and biophysical data, our structure is consistent with a model in which the two-helix finger and clamp cooperate during the ATPase cycle to move a polypeptide through the channel.
- State Key Laboratory of Membrane Biology, Peking-Tsinghua Center for Life Sciences, School of Life Sciences, Peking University, Beijing, China.
Organizational Affiliation: