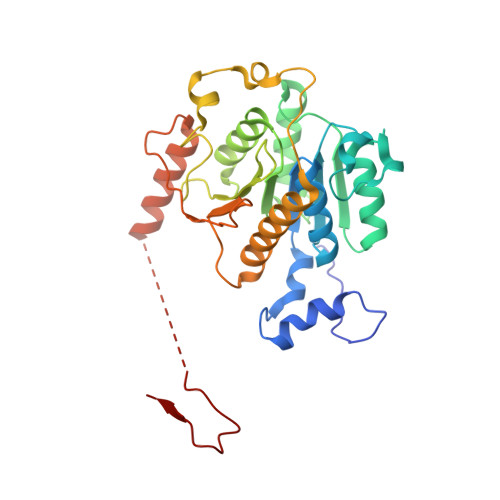
An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
Soudah, N., Padala, P., Hassouna, F., Kumar, M., Mashahreh, B., Lebedev, A.A., Isupov, M.N., Cohen-Kfir, E., Wiener, R.(2019) J Mol Biology 431: 463-478
- PubMed: 30412706
- DOI: https://doi.org/10.1016/j.jmb.2018.10.007
- Primary Citation of Related Structures:
6H77, 6H78 - PubMed Abstract:
Modification of proteins by the ubiquitin-like protein, UFM1, requires activation of UFM1 by the E1-activating enzyme, UBA5. In humans, UBA5 possesses two isoforms, each comprising an adenylation domain, but only one containing an N-terminal extension. Currently, the role of the N-terminal extension in UFM1 activation is not clear. Here we provide structural and biochemical data on UBA5 N-terminal extension to understand its contribution to UFM1 activation. The crystal structures of the UBA5 long isoform bound to ATP with and without UFM1 show that the N-terminus not only is directly involved in ATP binding but also affects how the adenylation domain interacts with ATP. Surprisingly, in the presence of the N-terminus, UBA5 no longer retains the 1:2 ratio of ATP to UBA5, but rather this becomes a 1:1 ratio. Accordingly, the N-terminus significantly increases the affinity of ATP to UBA5. Finally, the N-terminus, although not directly involved in the E2 binding, stimulates transfer of UFM1 from UBA5 to the E2, UFC1.
- Department of Biochemistry and Molecular Biology, The Institute for Medical Research Israel-Canada, Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel.
Organizational Affiliation: