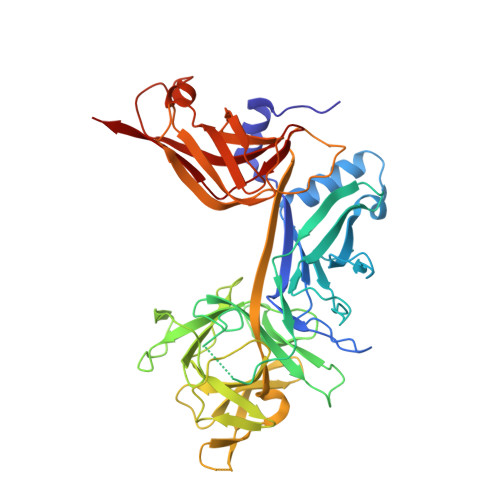
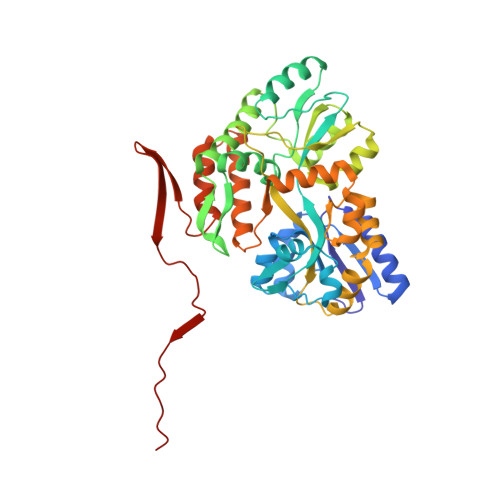
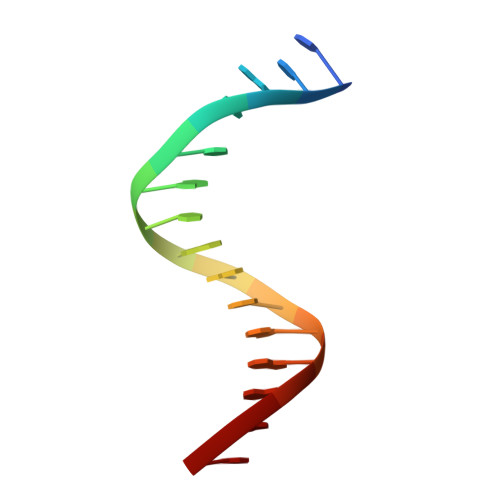
Structural and Functional Studies of the RBPJ-SHARP Complex Reveal a Conserved Corepressor Binding Site.
Yuan, Z., VanderWielen, B.D., Giaimo, B.D., Pan, L., Collins, C.E., Turkiewicz, A., Hein, K., Oswald, F., Borggrefe, T., Kovall, R.A.(2019) Cell Rep 26: 845-854.e6
- PubMed: 30673607
- DOI: https://doi.org/10.1016/j.celrep.2018.12.097
- Primary Citation of Related Structures:
6DKS - PubMed Abstract:
Notch is a conserved signaling pathway that is essential for metazoan development and homeostasis; dysregulated signaling underlies the pathophysiology of numerous human diseases. Receptor-ligand interactions result in gene expression changes, which are regulated by the transcription factor RBPJ. RBPJ forms a complex with the intracellular domain of the Notch receptor and the coactivator Mastermind to activate transcription, but it can also function as a repressor by interacting with corepressor proteins. Here, we determine the structure of RBPJ bound to the corepressor SHARP and DNA, revealing its mode of binding to RBPJ. We tested structure-based mutants in biophysical and biochemical-cellular assays to characterize the role of RBPJ as a repressor, clearly demonstrating that RBPJ mutants deficient for SHARP binding are incapable of repressing transcription of genes responsive to Notch signaling in cells. Altogether, our structure-function studies provide significant insights into the repressor function of RBPJ.
- Department of Molecular Genetics, Biochemistry and Microbiology, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Organizational Affiliation: