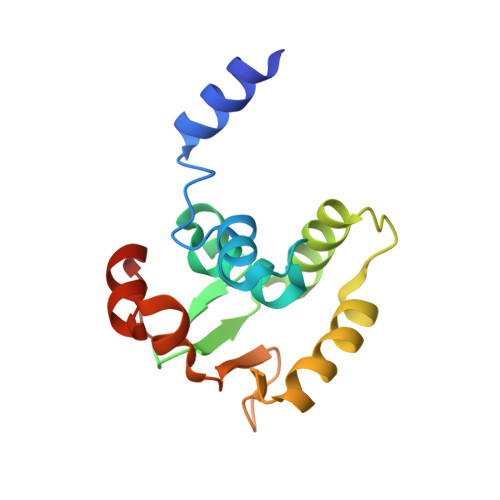
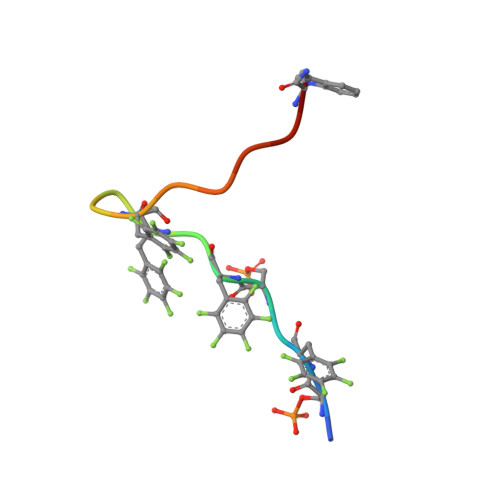
The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
Perez-Borrajero, C., Lin, C.S., Okon, M., Scheu, K., Graves, B.J., Murphy, M.E.P., McIntosh, L.P.(2019) J Mol Biology 431: 593-614
- PubMed: 30597162
- DOI: https://doi.org/10.1016/j.jmb.2018.12.011
- Primary Citation of Related Structures:
6DA1, 6DAT - PubMed Abstract:
The eukaryotic transcription factor ETS1 is regulated by an intrinsically disordered serine-rich region (SRR) that transiently associates with the adjacent ETS domain to inhibit DNA binding. In this study, we further elucidated the physicochemical basis for ETS1 autoinhibition by characterizing the interaction of its ETS domain with a series of synthetic peptides corresponding to the SRR. Binding is driven by the hydrophobic effect and enhanced electrostatically by phosphorylation of serines adjacent to aromatic residues in the amphipathic SRR. Structural characterization of the dynamic peptide/protein complex by NMR spectroscopy and X-ray crystallography revealed multiple modes of binding that lead to autoinhibition by synergistically blocking the DNA-binding interface of the ETS domain and stabilizing an appended helical inhibitory module against allosterically induced unfolding. Consistent with these conclusions, the SRR peptide does not interact with DNA-bound ETS1. In addition, we found that the ETS1 SRR phosphopeptide binds to distantly related PU.1 in vitro, indicating that autoinhibition exploits features of the ETS domain that are conserved across this family of transcription factors.
- Genome Sciences and Technology Program, University of British Columbia, Vancouver, BC, Canada V6T 1Z3; Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, BC, Canada V6T 1Z3.
Organizational Affiliation: