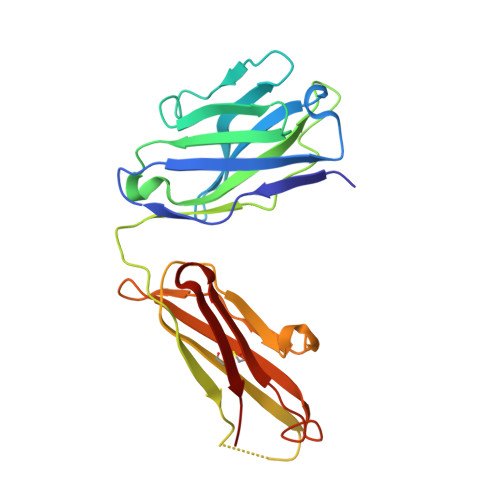
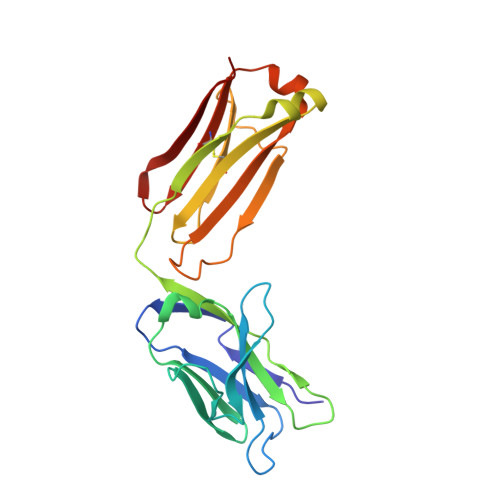
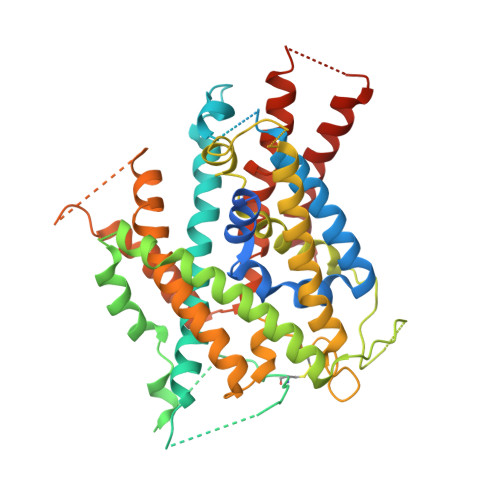
Crystal structure of arginine-bound lysosomal transporter SLC38A9 in the cytosol-open state.
Lei, H.T., Ma, J., Sanchez Martinez, S., Gonen, T.(2018) Nat Struct Mol Biol 25: 522-527
- PubMed: 29872228
- DOI: https://doi.org/10.1038/s41594-018-0072-2
- Primary Citation of Related Structures:
6C08 - PubMed Abstract:
Recent advances in understanding intracellular amino acid transport and mechanistic target of rapamycin complex 1 (mTORC1) signaling shed light on solute carrier 38, family A member 9 (SLC38A9), a lysosomal transporter responsible for the binding and translocation of several essential amino acids. Here we present the first crystal structure of SLC38A9 from Danio rerio in complex with arginine. As captured in the cytosol-open state, the bound arginine was locked in a transitional state stabilized by transmembrane helix 1 (TM1) of drSLC38A9, which was anchored at the groove between TM5 and TM7. These anchoring interactions were mediated by the highly conserved WNTMM motif in TM1, and mutations in this motif abolished arginine transport by drSLC38A9. The underlying mechanism of substrate binding is critical for sensitizing the mTORC1 signaling pathway to amino acids and for maintenance of lysosomal amino acid homeostasis. This study offers a first glimpse into a prototypical model for SLC38 transporters.
- Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA, USA.
Organizational Affiliation: