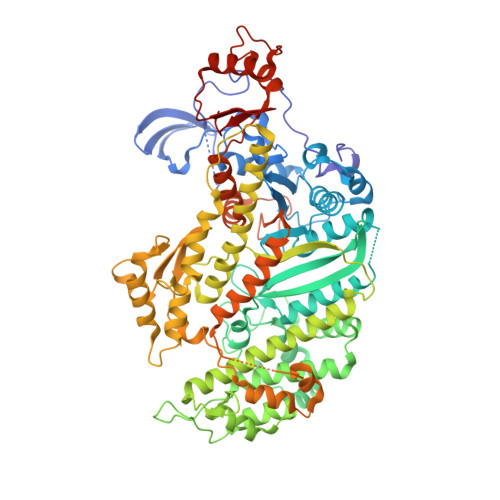
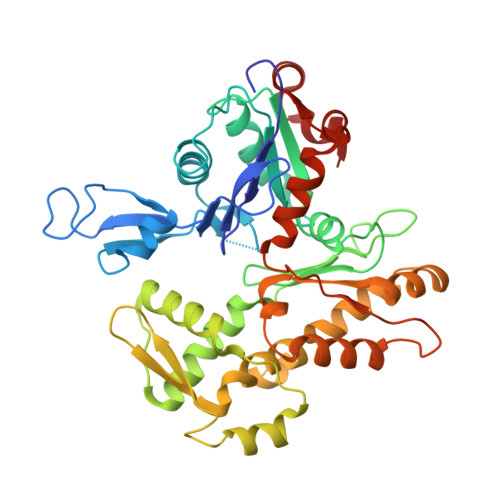
The structure of the actin-smooth muscle myosin motor domain complex in the rigor state.
Banerjee, C., Hu, Z., Huang, Z., Warrington, J.A., Taylor, D.W., Trybus, K.M., Lowey, S., Taylor, K.A.(2017) J Struct Biol 200: 325-333
- PubMed: 29038012
- DOI: https://doi.org/10.1016/j.jsb.2017.10.003
- Primary Citation of Related Structures:
6BIH - PubMed Abstract:
Myosin-based motility utilizes catalysis of ATP to drive the relative sliding of F-actin and myosin. The earliest detailed model based on cryo-electron microscopy (cryoEM) and X-ray crystallography postulated that higher actin affinity and lever arm movement were coupled to closure of a feature of the myosin head dubbed the actin-binding cleft. Several studies since then using crystallography of myosin-V and cryoEM structures of F-actin bound myosin-I, -II and -V have provided details of this model. The smooth muscle myosin II interaction with F-actin may differ from those for striated and non-muscle myosin II due in part to different lengths of important surface loops. Here we report a ∼6 Å resolution reconstruction of F-actin decorated with the nucleotide-free recombinant smooth muscle myosin-II motor domain (MD) from images recorded using a direct electron detector. Resolution is highest for F-actin and the actin-myosin interface (3.5-4 Å) and lowest (∼6-7 Å) for those parts of the MD at the highest radius. Atomic models built into the F-actin density are quite comparable to those previously reported for rabbit muscle actin and show density from the bound ADP. The atomic model of the MD, is quite similar to a recently published structure of vertebrate non-muscle myosin II bound to F-actin and a crystal structure of nucleotide free myosin-V. Larger differences are observed when compared to the cryoEM structure of F-actin decorated with rabbit skeletal muscle myosin subfragment 1. The differences suggest less closure of the 50 kDa domain in the actin bound skeletal muscle myosin structure.
- Department of Computer Science, Florida State University, Tallahassee, FL 32306-4530, United States.
Organizational Affiliation: