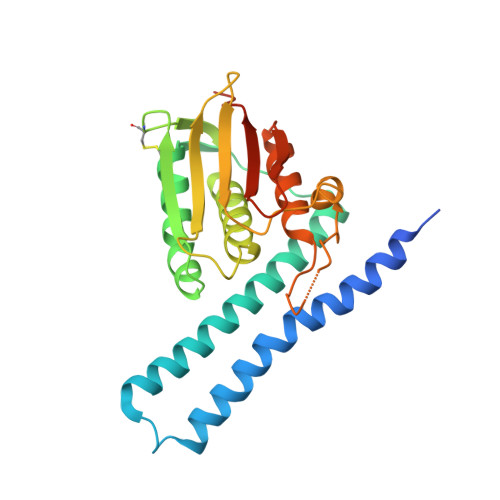
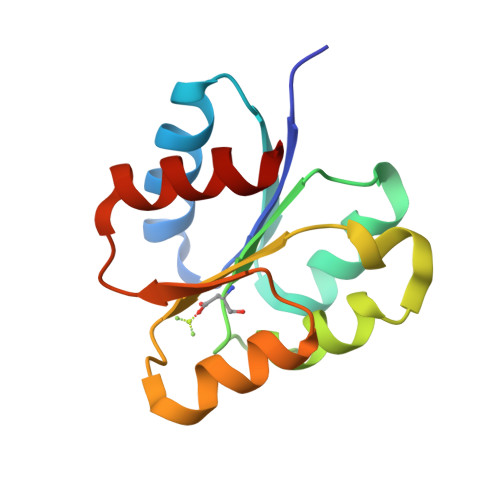
A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Liu, Y., Rose, J., Huang, S., Hu, Y., Wu, Q., Wang, D., Li, C., Liu, M., Zhou, P., Jiang, L.(2017) Nat Commun 8: 2104-2104
- PubMed: 29235472
- DOI: https://doi.org/10.1038/s41467-017-02310-9
- Primary Citation of Related Structures:
5UHT, 6AZR - PubMed Abstract:
Histidine kinases are key regulators in the bacterial two-component systems that mediate the cellular response to environmental changes. The vast majority of the sensor histidine kinases belong to the bifunctional HisKA family, displaying both kinase and phosphatase activities toward their substrates. The molecular mechanisms regulating the opposing activities of these enzymes are not well understood. Through a combined NMR and crystallographic study on the histidine kinase HK853 and its response regulator RR468 from Thermotoga maritima, here we report a pH-mediated conformational switch of HK853 that shuts off its phosphatase activity under acidic conditions. Such a pH-sensing mechanism is further demonstrated in the EnvZ-OmpR two-component system from Salmonella enterica in vitro and in vivo, which directly contributes to the bacterial infectivity. Our finding reveals a broadly conserved mechanism that regulates the phosphatase activity of the largest family of bifunctional histidine kinases in response to the change of environmental pH.
- Key Laboratory of Magnetic Resonance in Biological Systems, State Key Laboratory of Magnetic Resonance and Atomic and Molecular Physics, National Center for Magnetic Resonance in Wuhan, Wuhan Institute of Physics and Mathematics, Chinese Academy of Sciences, Wuhan, 430071, China.
Organizational Affiliation: