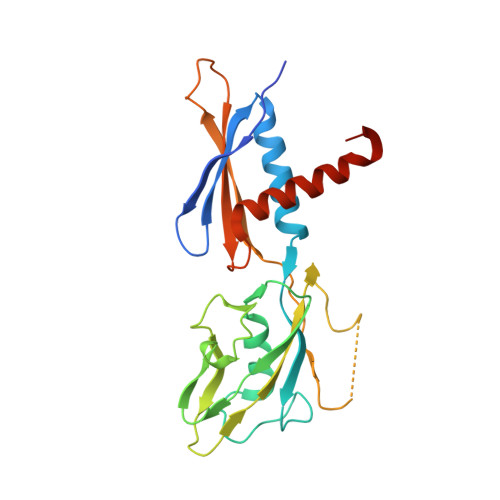
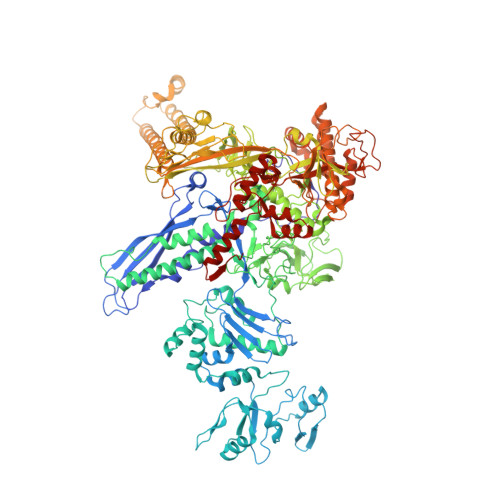
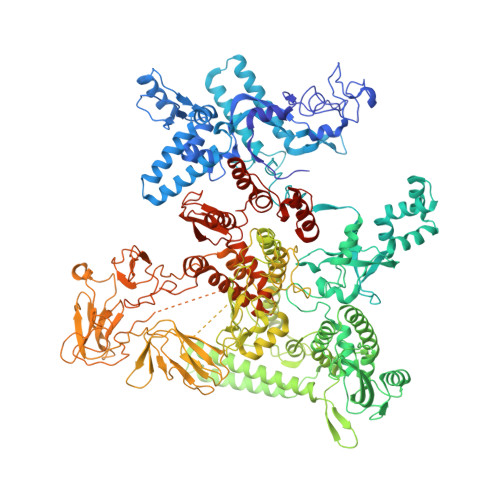
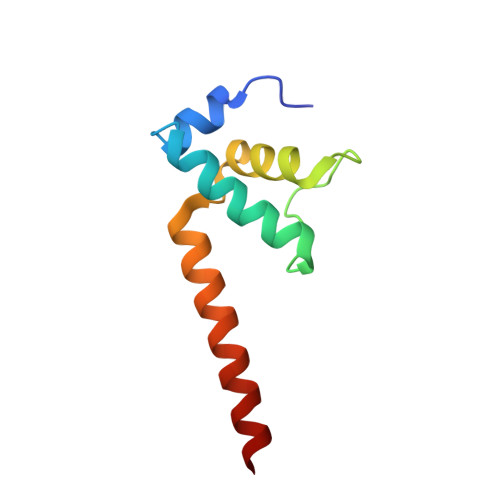
RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing.
Kang, J.Y., Mishanina, T.V., Bellecourt, M.J., Mooney, R.A., Darst, S.A., Landick, R.(2018) Mol Cell 69: 802-815.e1
- PubMed: 29499135
- DOI: https://doi.org/10.1016/j.molcel.2018.01.018
- Primary Citation of Related Structures:
6ASX, 6BJS - PubMed Abstract:
Sequence-specific pausing by RNA polymerase (RNAP) during transcription plays crucial and diverse roles in gene expression. In bacteria, RNA structures are thought to fold within the RNA exit channel of the RNAP and can increase pause lifetimes significantly. The biophysical mechanism of pausing is uncertain. We used single-particle cryo-EM to determine structures of paused complexes, including a 3.8-Å structure of an RNA hairpin-stabilized, paused RNAP that coordinates RNA folding in the his operon attenuation control region of E. coli. The structures revealed a half-translocated pause state (RNA post-translocated, DNA pre-translocated) that can explain transcriptional pausing and a global conformational change of RNAP that allosterically inhibits trigger loop folding and can explain pause hairpin action. Pause hairpin interactions with the RNAP RNA exit channel suggest how RNAP guides the formation of nascent RNA structures.
- The Rockefeller University, 1230 York Avenue, New York, NY 10065, USA.
Organizational Affiliation: