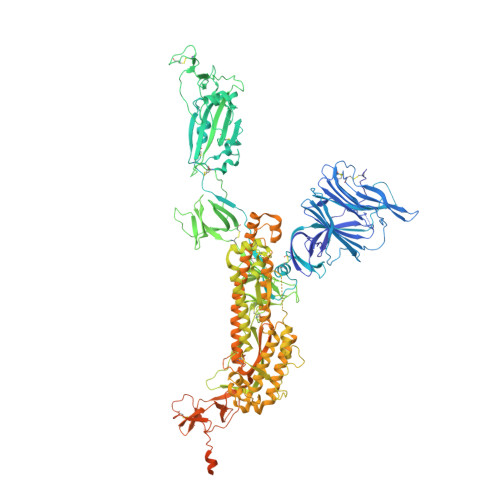
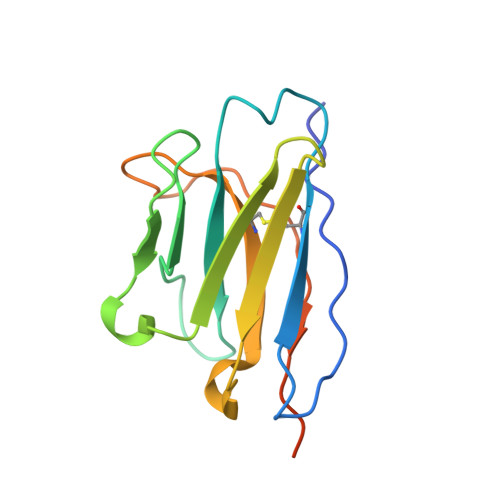
An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction.
Hanke, L., Vidakovics Perez, L., Sheward, D.J., Das, H., Schulte, T., Moliner-Morro, A., Corcoran, M., Achour, A., Karlsson Hedestam, G.B., Hallberg, B.M., Murrell, B., McInerney, G.M.(2020) Nat Commun 11: 4420-4420
- PubMed: 32887876
- DOI: https://doi.org/10.1038/s41467-020-18174-5
- Primary Citation of Related Structures:
6ZXN - PubMed Abstract:
SARS-CoV-2 enters host cells through an interaction between the spike glycoprotein and the angiotensin converting enzyme 2 (ACE2) receptor. Directly preventing this interaction presents an attractive possibility for suppressing SARS-CoV-2 replication. Here, we report the isolation and characterization of an alpaca-derived single domain antibody fragment, Ty1, that specifically targets the receptor binding domain (RBD) of the SARS-CoV-2 spike, directly preventing ACE2 engagement. Ty1 binds the RBD with high affinity, occluding ACE2. A cryo-electron microscopy structure of the bound complex at 2.9 Å resolution reveals that Ty1 binds to an epitope on the RBD accessible in both the 'up' and 'down' conformations, sterically hindering RBD-ACE2 binding. While fusion to an Fc domain renders Ty1 extremely potent, Ty1 neutralizes SARS-CoV-2 spike pseudovirus as a 12.8 kDa nanobody, which can be expressed in high quantities in bacteria, presenting opportunities for manufacturing at scale. Ty1 is therefore an excellent candidate as an intervention against COVID-19.
- Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet, Stockholm, Sweden.
Organizational Affiliation: