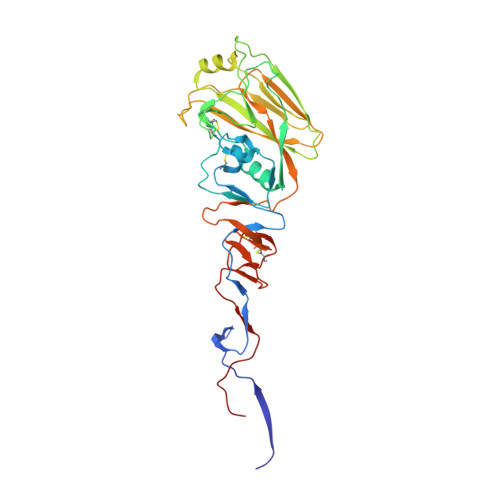
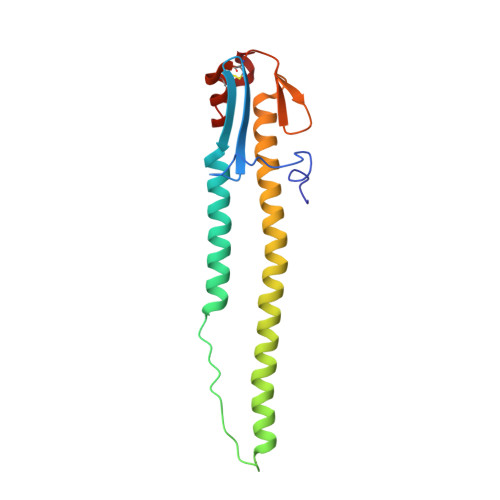
Hemagglutinin Structure and Activities.
Gamblin, S.J., Vachieri, S.G., Xiong, X., Zhang, J., Martin, S.R., Skehel, J.J.(2021) Cold Spring Harb Perspect Med 11
- PubMed: 32513673
- DOI: https://doi.org/10.1101/cshperspect.a038638
- Primary Citation of Related Structures:
6ZRK, 7A9D - PubMed Abstract:
Hemagglutinins (HAs) are the receptor-binding and membrane fusion glycoproteins of influenza viruses. They recognize sialic acid-containing, cell-surface glycoconjugates as receptors but have limited affinity for them, and, as a consequence, virus attachment to cells requires their interaction with several virus HAs. Receptor-bound virus is transferred into endosomes where membrane fusion by HAs is activated at pH between 5 and 6.5, depending on the strain of virus. Fusion activity requires extensive rearrangements in HA conformation that include extrusion of a buried "fusion peptide" to connect with the endosomal membrane, form a bridge to the virus membrane, and eventually bring both membranes close together. In this review, we give an overview of the structures of the 16 genetically and antigenically distinct subtypes of influenza A HA in relation to these two functions in virus replication and in relation to recognition of HA by antibodies that neutralize infection.
- Structural Biology of Disease Processes Laboratory, The Francis Crick Institute, London NW1 1AT, United Kingdom.
Organizational Affiliation: