Insulin binding to the analytical antibody sandwich pair OXI-005 and HUI-018: Epitope mapping and binding properties.
Johansson, E., Wu, X., Yu, B., Yang, Z., Cao, Z., Wiberg, C., Jeppesen, C.B., Poulsen, F.(2021) Protein Sci 30: 485-496
- PubMed: 33277949
- DOI: https://doi.org/10.1002/pro.4009
- Primary Citation of Related Structures:
6Z7W, 6Z7X, 6Z7Y, 6Z7Z - PubMed Abstract:
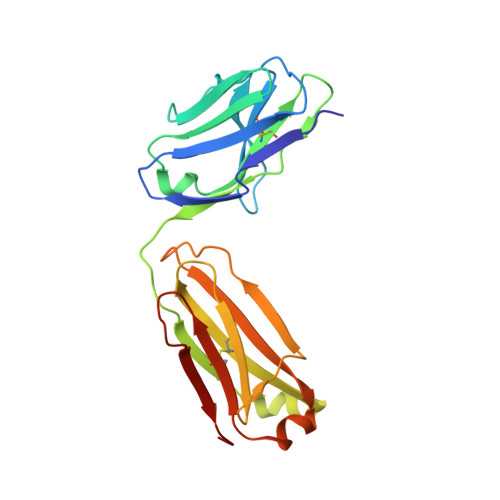
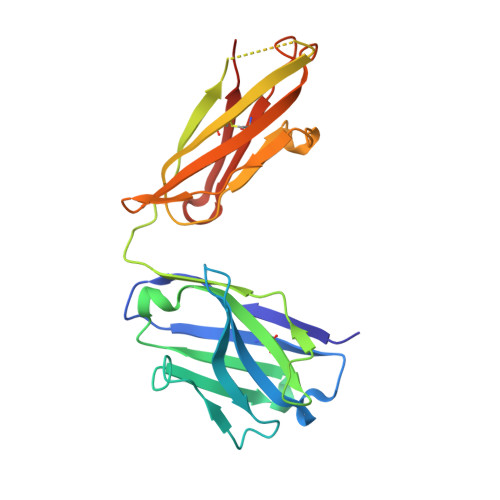
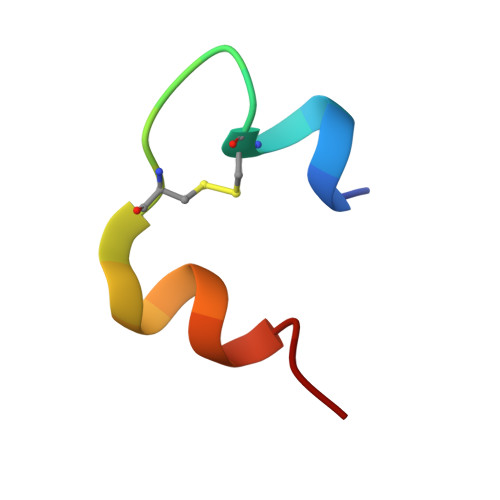
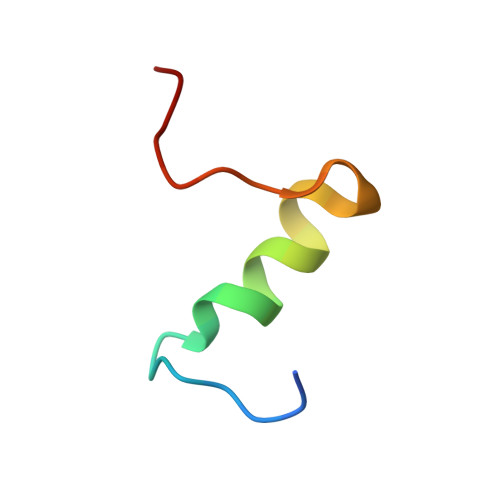
The insulin epitopes for two monoclonal antibodies (mAbs), OXI-005 and HUI-018, commonly used in combination for insulin concentration determination in sandwich assays, were determined using X-ray crystallography. The crystal structure of the HUI-018 Fab in complex with human insulin (HI) was determined and OXI-005 Fab crystal structures were determined in complex with HI and porcine insulin (PI) as well as on its own. The OXI-005 epitope comprises insulin residues 1,3,4,19-21 (A-chain) and 25-30 (B-chain) and for HUI-018 residues 7,8,10-14,17 (A-chain) and 5-7, 10, 14 (B-chain). The areas of insulin involved in interactions with the mAb are 20% (OXI-005) and 24% (HUI-018) of the total insulin surface. Based on the Fab complex crystal structures with the insulins a molecular model for simultaneous binding of the Fabs to PI was built and this model was validated by small angle X-ray scattering measurements for the ternary complex. The epitopes for the mAbs on insulin were found well separated from each other as expected from luminiscent oxygen channeling immunoassay results for different insulins (HI, PI, bovine insulin, DesB30 HI, insulin glargine, insulin lispro). The affinities of the OXI-005 and HUI-018 Fabs for HI, PI, and DesB30 HI were determined using surface plasmon resonance. The K D s were found to be in the range of 1-4 nM for the HUI-018 Fab, while more different for the OXI-005 Fab (50 nM for HI, 20 nM for PI and 400 nM for DesB30 HI) supporting the importance of residue B30 for binding to OXI-005.
- Novo Nordisk A/S, Måløv, Denmark.
Organizational Affiliation: