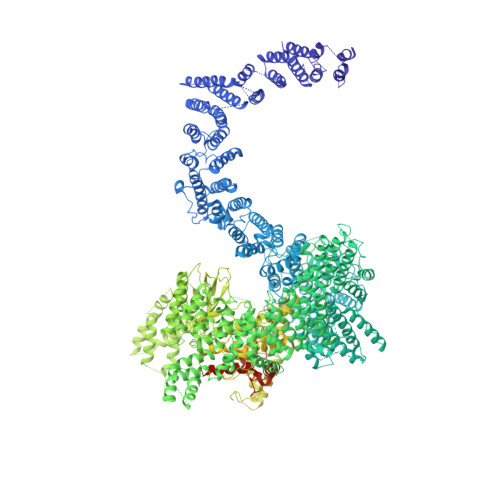
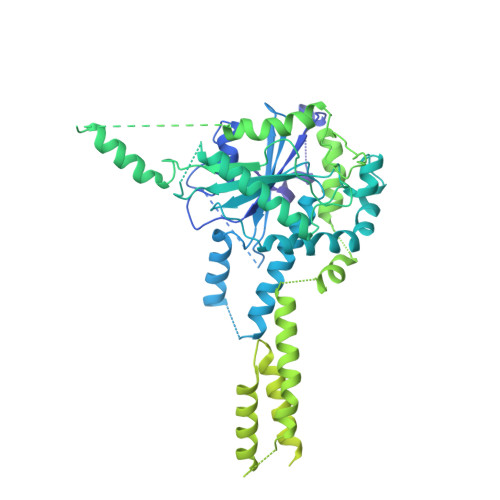
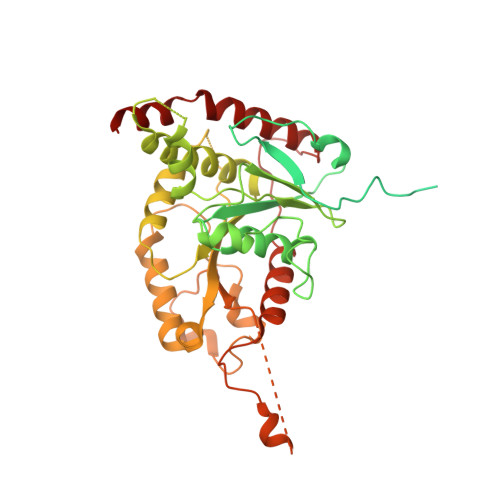
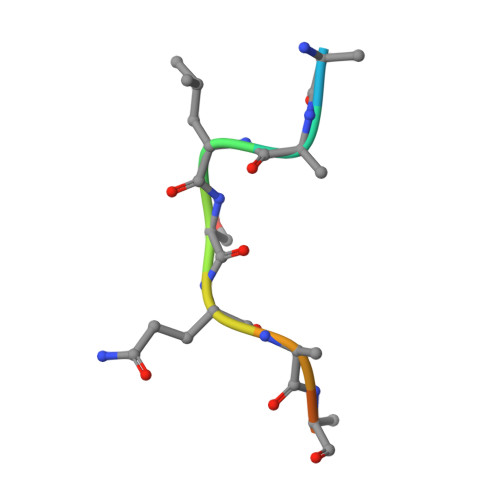
Structure of substrate-bound SMG1-8-9 kinase complex reveals molecular basis for phosphorylation specificity.
Langer, L.M., Gat, Y., Bonneau, F., Conti, E.(2020) Elife 9
- PubMed: 32469312
- DOI: https://doi.org/10.7554/eLife.57127
- Primary Citation of Related Structures:
6Z3R - PubMed Abstract:
PI3K-related kinases (PIKKs) are large Serine/Threonine (Ser/Thr)-protein kinases central to the regulation of many fundamental cellular processes. PIKK family member SMG1 orchestrates progression of an RNA quality control pathway, termed nonsense-mediated mRNA decay (NMD), by phosphorylating the NMD factor UPF1. Phosphorylation of UPF1 occurs in its unstructured N- and C-terminal regions at Serine/Threonine-Glutamine (SQ) motifs. How SMG1 and other PIKKs specifically recognize SQ motifs has remained unclear. Here, we present a cryo-electron microscopy (cryo-EM) reconstruction of a human SMG1-8-9 kinase complex bound to a UPF1 phosphorylation site at an overall resolution of 2.9 Å. This structure provides the first snapshot of a human PIKK with a substrate-bound active site. Together with biochemical assays, it rationalizes how SMG1 and perhaps other PIKKs specifically phosphorylate Ser/Thr-containing motifs with a glutamine residue at position +1 and a hydrophobic residue at position -1, thus elucidating the molecular basis for phosphorylation site recognition.
- Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Martinsried, Germany.
Organizational Affiliation: