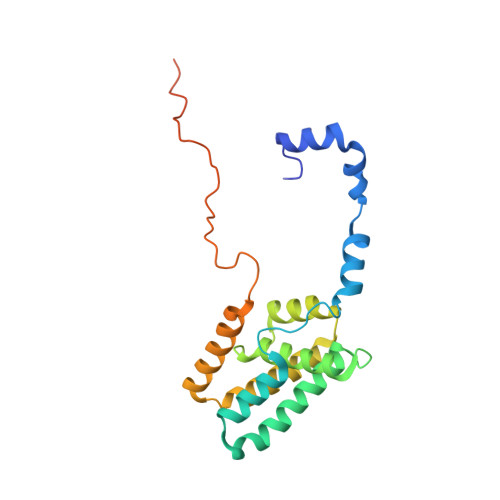
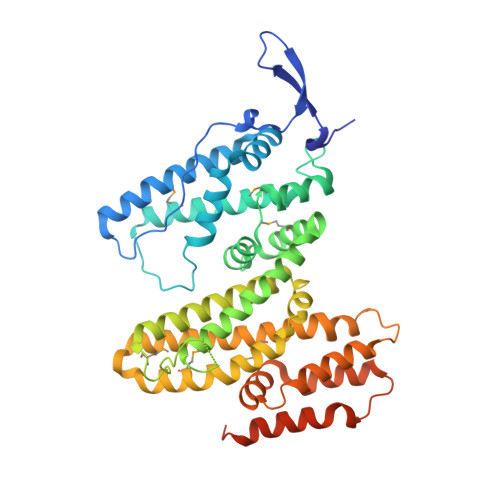
The interaction of DNA repair factors ASCC2 and ASCC3 is affected by somatic cancer mutations.
Jia, J., Absmeier, E., Holton, N., Pietrzyk-Brzezinska, A.J., Hackert, P., Bohnsack, K.E., Bohnsack, M.T., Wahl, M.C.(2020) Nat Commun 11: 5535-5535
- PubMed: 33139697
- DOI: https://doi.org/10.1038/s41467-020-19221-x
- Primary Citation of Related Structures:
6YXQ - PubMed Abstract:
The ASCC3 subunit of the activating signal co-integrator complex is a dual-cassette Ski2-like nucleic acid helicase that provides single-stranded DNA for alkylation damage repair by the α-ketoglutarate-dependent dioxygenase AlkBH3. Other ASCC components integrate ASCC3/AlkBH3 into a complex DNA repair pathway. We mapped and structurally analyzed interacting ASCC2 and ASCC3 regions. The ASCC3 fragment comprises a central helical domain and terminal, extended arms that clasp the compact ASCC2 unit. ASCC2-ASCC3 interfaces are evolutionarily highly conserved and comprise a large number of residues affected by somatic cancer mutations. We quantified contributions of protein regions to the ASCC2-ASCC3 interaction, observing that changes found in cancers lead to reduced ASCC2-ASCC3 affinity. Functional dissection of ASCC3 revealed similar organization and regulation as in the spliceosomal RNA helicase Brr2. Our results delineate functional regions in an important DNA repair complex and suggest possible molecular disease principles.
- Laboratory of Structural Biochemistry, Freie Universität Berlin, D-14195, Berlin, Germany.
Organizational Affiliation: