The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13
Patel, H.M., Roszak, A.W., Madamwar, D., Cogdell, R.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Allophycocyanin alpha | 160 | Nostoc sp. WR13 | Mutation(s): 0 | 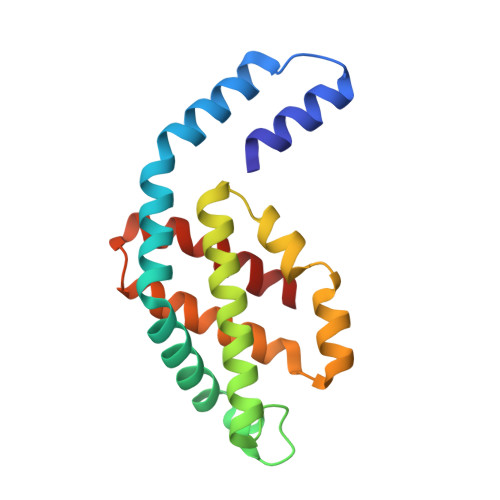 | |
UniProt | |||||
Find proteins for A0A4Y5PW22 (Nostoc sp. WR13) Explore A0A4Y5PW22 Go to UniProtKB: A0A4Y5PW22 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A4Y5PW22 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Allophycocyanin beta | 161 | Nostoc sp. WR13 | Mutation(s): 0 | 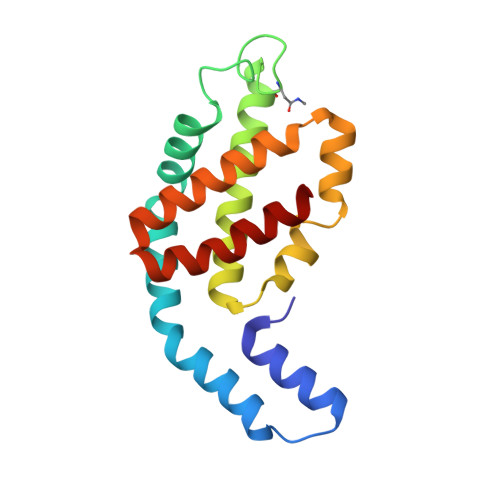 | |
UniProt | |||||
Find proteins for A0A4Y5PW23 (Nostoc sp. WR13) Explore A0A4Y5PW23 Go to UniProtKB: A0A4Y5PW23 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A4Y5PW23 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 14 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CYC (Subject of Investigation/LOI) Query on CYC | CA [auth DDD] CB [auth KKK] FA [auth EEE] GB [auth LLL] IA [auth FFF] | PHYCOCYANOBILIN C33 H40 N4 O6 VXTXPYZGDQPMHK-GMXXPEQVSA-N |  | ||
| P6G Query on P6G | X [auth CCC] | HEXAETHYLENE GLYCOL C12 H26 O7 IIRDTKBZINWQAW-UHFFFAOYSA-N |  | ||
| 1PE Query on 1PE | R [auth BBB] | PENTAETHYLENE GLYCOL C10 H22 O6 JLFNLZLINWHATN-UHFFFAOYSA-N |  | ||
| PG4 Query on PG4 | AB [auth JJJ], LA [auth FFF], N [auth AAA], Y [auth CCC] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| PGE Query on PGE | JB [auth LLL], OA [auth GGG], PA [auth GGG] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| LYS Query on LYS | DB [auth KKK], P [auth BBB], WA [auth III] | LYSINE C6 H15 N2 O2 KDXKERNSBIXSRK-YFKPBYRVSA-O |  | ||
| GLU Query on GLU | DA [auth DDD], SA [auth HHH] | GLUTAMIC ACID C5 H9 N O4 WHUUTDBJXJRKMK-VKHMYHEASA-N |  | ||
| MRD Query on MRD | NA [auth GGG] | (4R)-2-METHYLPENTANE-2,4-DIOL C6 H14 O2 SVTBMSDMJJWYQN-RXMQYKEDSA-N |  | ||
| MPD Query on MPD | EA [auth DDD] HA [auth EEE] IB [auth LLL] KA [auth FFF] Q [auth BBB] | (4S)-2-METHYL-2,4-PENTANEDIOL C6 H14 O2 SVTBMSDMJJWYQN-YFKPBYRVSA-N |  | ||
| PEG Query on PEG | BB [auth JJJ] QA [auth GGG] S [auth BBB] T [auth BBB] UA [auth HHH] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| DSN Query on DSN | HB [auth LLL], YA [auth JJJ] | D-SERINE C3 H7 N O3 MTCFGRXMJLQNBG-UWTATZPHSA-N |  | ||
| ALA Query on ALA | JA [auth FFF] | ALANINE C3 H7 N O2 QNAYBMKLOCPYGJ-REOHCLBHSA-N |  | ||
| GLY Query on GLY | GA [auth EEE] | GLYCINE C2 H5 N O2 DHMQDGOQFOQNFH-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | AA [auth CCC], BA [auth CCC], EB [auth KKK], FB [auth KKK], U [auth BBB] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MEN Query on MEN | B [auth BBB] D [auth DDD] F [auth FFF] H [auth HHH] J [auth JJJ] B [auth BBB], D [auth DDD], F [auth FFF], H [auth HHH], J [auth JJJ], L [auth LLL] | L-PEPTIDE LINKING | C5 H10 N2 O3 |  | ASN |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 123.695 | α = 90 |
| b = 177.726 | β = 90 |
| c = 106.372 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| autoPROC | data scaling |
| STARANISO | data scaling |
| PHASER | phasing |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Department of Energy (DOE, United States) | United States | DE-SC0001035 (Office of Basic Energy Sciences) |
| Department of Biotechnology (DBT, India) | India | BT/PR15686/AAQ/3/811/2016 (UGC-BSR Faculty Fellowship) |
| Department of Biotechnology (DBT, India) | India | BT/IN/UK/DBT-BC/2017-18 (Newton-Bhabha fellowship) |