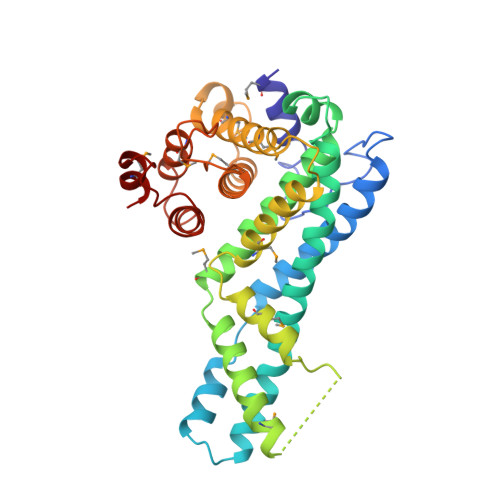
Structure of CYRI-B (FAM49B), a key regulator of cellular actin assembly.
Kaplan, E., Stone, R., Hume, P.J., Greene, N.P., Koronakis, V.(2020) Acta Crystallogr D Struct Biol 76: 1015-1024
- PubMed: 33021503
- DOI: https://doi.org/10.1107/S2059798320010906
- Primary Citation of Related Structures:
6YJJ, 6YJK - PubMed Abstract:
In eukaryotes, numerous fundamental processes are controlled by the WAVE regulatory complex (WRC) that regulates cellular actin polymerization, crucial for cell motility, cell-cell adhesion and epithelial differentiation. Actin assembly is triggered by interaction of the small GTPase Rac1 with CYFIP1, a key component of the WRC. Previously known as FAM49B, CYRI-B is a protein that is highly conserved across the Eukaryota and has recently been revealed to be a key regulator of Rac1 activity. Mutation of CYRI-B or alteration of its expression therefore leads to altered actin nucleation dynamics, with impacts on lamellipodia formation, cell migration and infection by intracellular pathogens. In addition, knockdown of CYRI-B expression in cancer cell lines results in accelerated cell proliferation and invasiveness. Here, the structure of Rhincodon typus (whale shark) CYRI-B is presented, which is the first to be reported of any CYRI family member. Solved by X-ray crystallography, the structure reveals that CYRI-B comprises three distinct α-helical subdomains and is highly structurally related to a conserved domain present in CYFIP proteins. The work presented here establishes a template towards a better understanding of CYRI-B biological function.
- Department of Pathology, University of Cambridge, Tennis Court Road, Cambridge CB2 1QP, United Kingdom.
Organizational Affiliation: