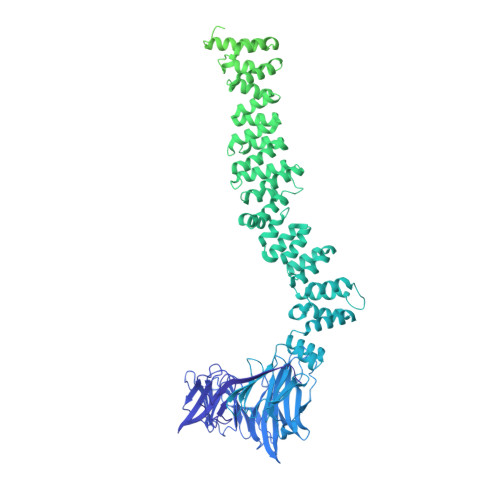
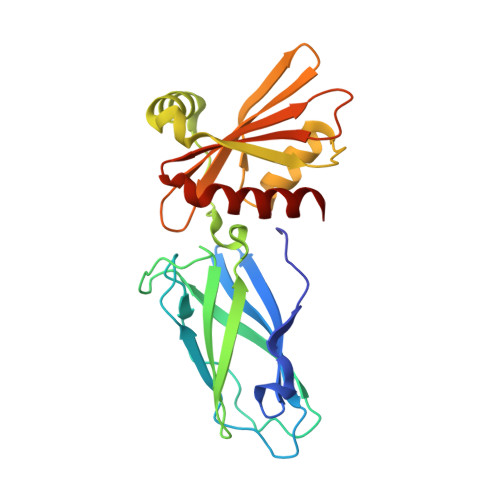
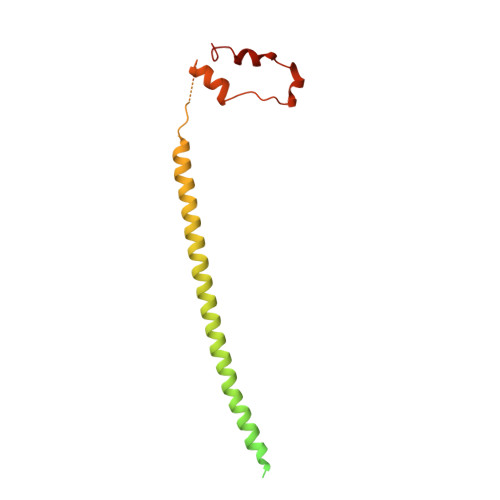
Architecture of the AP2/clathrin coat on the membranes of clathrin-coated vesicles.
Kovtun, O., Dickson, V.K., Kelly, B.T., Owen, D.J., Briggs, J.A.G.(2020) Sci Adv 6: eaba8381-eaba8381
- PubMed: 32743075
- DOI: https://doi.org/10.1126/sciadv.aba8381
- Primary Citation of Related Structures:
6YAE, 6YAF, 6YAH, 6YAI - PubMed Abstract:
Clathrin-mediated endocytosis (CME) is crucial for modulating the protein composition of a cell's plasma membrane. Clathrin forms a cage-like, polyhedral outer scaffold around a vesicle, to which cargo-selecting clathrin adaptors are attached. Adaptor protein complex (AP2) is the key adaptor in CME. Crystallography has shown AP2 to adopt a range of conformations. Here, we used cryo-electron microscopy, tomography, and subtomogram averaging to determine structures, interactions, and arrangements of clathrin and AP2 at the key steps of coat assembly, from AP2 in solution to membrane-assembled clathrin-coated vesicles (CCVs). AP2 binds cargo and PtdIns(4,5) P 2 (phosphatidylinositol 4,5-bisphosphate)-containing membranes via multiple interfaces, undergoing conformational rearrangement from its cytosolic state. The binding mode of AP2 β2 appendage into the clathrin lattice in CCVs and buds implies how the adaptor structurally modulates coat curvature and coat disassembly.
- MRC Laboratory of Molecular Biology, Cambridge Biomedical Campus, Cambridge CB2 0QH, UK.
Organizational Affiliation: