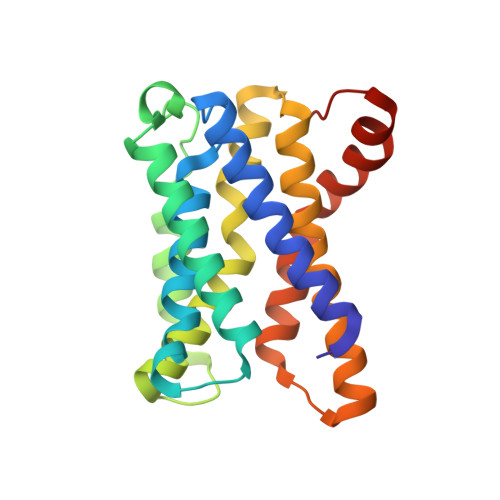
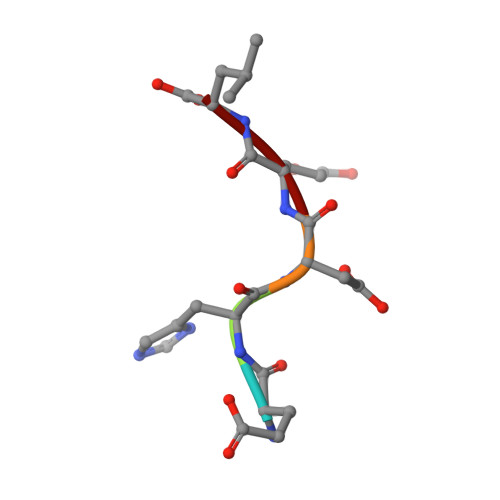
A signal capture and proofreading mechanism for the KDEL-receptor explains selectivity and dynamic range in ER retrieval.
Gerondopoulos, A., Brauer, P., Sobajima, T., Wu, Z., Parker, J.L., Biggin, P.C., Barr, F.A., Newstead, S.(2021) Elife 10
- PubMed: 34137369
- DOI: https://doi.org/10.7554/eLife.68380
- Primary Citation of Related Structures:
6Y7V, 6ZXR - PubMed Abstract:
ER proteins of widely differing abundance are retrieved from the Golgi by the KDEL-receptor. Abundant ER proteins tend to have KDEL rather than HDEL signals, whereas ADEL and DDEL are not used in most organisms. Here, we explore the mechanism of selective retrieval signal capture by the KDEL-receptor and how HDEL binds with 10-fold higher affinity than KDEL. Our results show the carboxyl-terminus of the retrieval signal moves along a ladder of arginine residues as it enters the binding pocket of the receptor. Gatekeeper residues D50 and E117 at the entrance of this pocket exclude ADEL and DDEL sequences. D50N/E117Q mutation of human KDEL-receptors changes the selectivity to ADEL and DDEL. However, further analysis of HDEL, KDEL, and RDEL-bound receptor structures shows that affinity differences are explained by interactions between the variable -4 H/K/R position of the signal and W120, rather than D50 or E117. Together, these findings explain KDEL-receptor selectivity, and how signal variants increase dynamic range to support efficient ER retrieval of low and high abundance proteins.
- Department of Biochemistry, University of Oxford, Oxford, United Kingdom.
Organizational Affiliation: