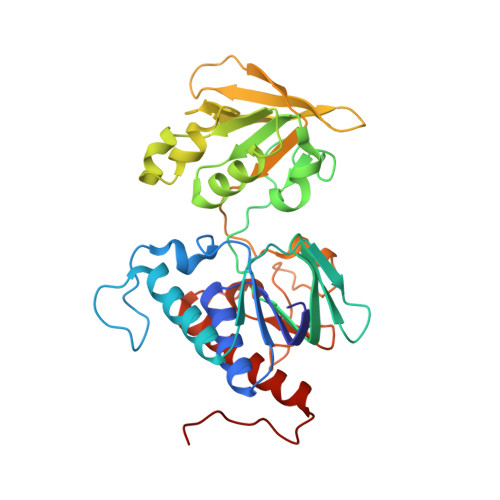
Unexpected diversity of ferredoxin-dependent thioredoxin reductases in cyanobacteria.
Buey, R.M., Fernandez-Justel, D., Gonzalez-Holgado, G., Martinez-Julvez, M., Gonzalez-Lopez, A., Velazquez-Campoy, A., Medina, M., Buchanan, B.B., Balsera, M.(2021) Plant Physiol 186: 285-296
- PubMed: 33599267
- DOI: https://doi.org/10.1093/plphys/kiab072
- Primary Citation of Related Structures:
6XTF - PubMed Abstract:
Thioredoxin reductases control the redox state of thioredoxins (Trxs)-ubiquitous proteins that regulate a spectrum of enzymes by dithiol-disulfide exchange reactions. In most organisms, Trx is reduced by NADPH via a thioredoxin reductase flavoenzyme (NTR), but in oxygenic photosynthetic organisms, this function can also be performed by an iron-sulfur ferredoxin (Fdx)-dependent thioredoxin reductase (FTR) that links light to metabolic regulation. We have recently found that some cyanobacteria, such as the thylakoid-less Gloeobacter and the ocean-dwelling green oxyphotobacterium Prochlorococcus, lack NTR and FTR but contain a thioredoxin reductase flavoenzyme (formerly tentatively called deeply-rooted thioredoxin reductase or DTR), whose electron donor remained undefined. Here, we demonstrate that Fdx functions in this capacity and report the crystallographic structure of the transient complex between the plant-type Fdx1 and the thioredoxin reductase flavoenzyme from Gloeobacter violaceus. Thereby, our data demonstrate that this cyanobacterial enzyme belongs to the Fdx flavin-thioredoxin reductase (FFTR) family, originally described in the anaerobic bacterium Clostridium pasteurianum. Accordingly, the enzyme hitherto termed DTR is renamed FFTR. Our experiments further show that the redox-sensitive peptide CP12 is modulated in vitro by the FFTR/Trx system, demonstrating that FFTR functionally substitutes for FTR in light-linked enzyme regulation in Gloeobacter. Altogether, we demonstrate the FFTR is spread within the cyanobacteria phylum and propose that, by substituting for FTR, it connects the reduction of target proteins to photosynthesis. Besides, the results indicate that FFTR acquisition constitutes a mechanism of evolutionary adaptation in marine phytoplankton such as Prochlorococcus that live in low-iron environments.
- Metabolic Engineering Group, Departamento de Microbiología y Genética, Universidad de Salamanca, Salamanca 37007, Spain.
Organizational Affiliation: