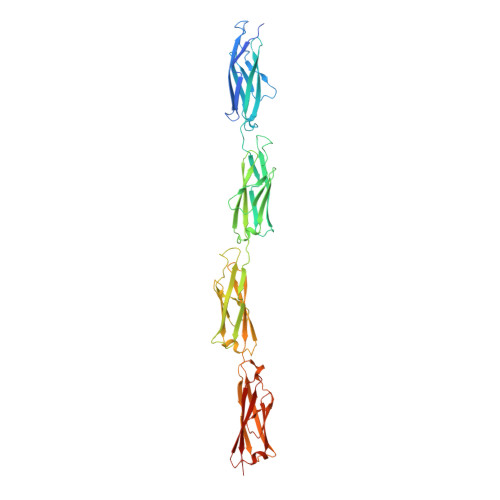Essential role of calcium in extending RTX adhesins to their target.
Vance, T.D.R., Ye, Q., Conroy, B., Davies, P.L.(2020) J Struct Biol X 4: 100036-100036
- PubMed: 32984811
- DOI: https://doi.org/10.1016/j.yjsbx.2020.100036
- Primary Citation of Related Structures:
6XI1, 6XI3 - PubMed Abstract:
RTX adhesins are long, multi-domain proteins present on the outer membrane of many Gram-negative bacteria. From this vantage point, adhesins use their distal ligand-binding domains for surface attachment leading to biofilm formation. To expand the reach of the ligand-binding domains, RTX adhesins maintain a central extender region of multiple tandem repeats, which makes up most of the proteins' large molecular weight. Alignments of the 10-15-kDa extender domains show low sequence identity between adhesins. Here we have produced and structurally characterized protein constructs of four tandem repeats (tetra-tandemers) from two different RTX adhesins. In comparing the tetra-tandemers to each other and already solved structures from Marinomonas primoryensis and Salmonella enterica , the extender domains fold as diverse beta-sandwich structures with widely differing calcium contents. However, all the tetra-tandemers have at least one calcium ion coordinated in the linker region between beta-sandwich domains whose role appears to be the rigidification of the extender region to help the adhesin extend its reach.
- Department of Biomedical and Molecular Science, Queen's University Kingston ON, Canada.
Organizational Affiliation: