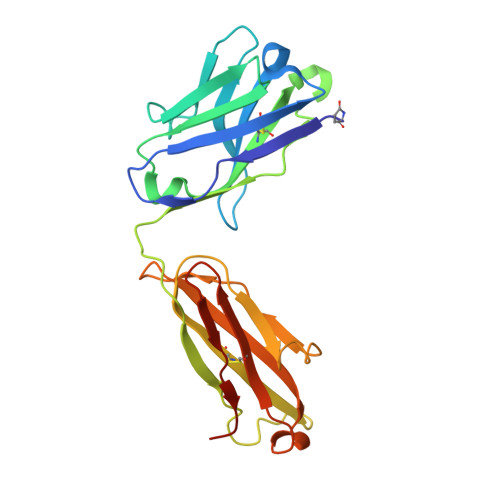
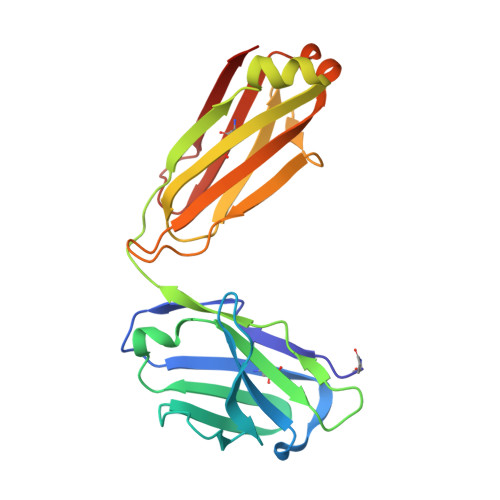
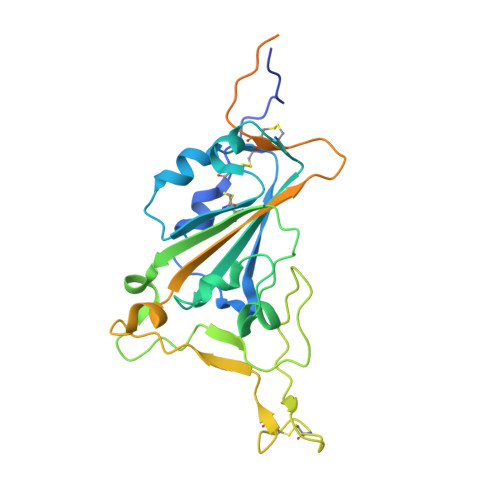
Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation.
Hurlburt, N.K., Seydoux, E., Wan, Y.H., Edara, V.V., Stuart, A.B., Feng, J., Suthar, M.S., McGuire, A.T., Stamatatos, L., Pancera, M.(2020) Nat Commun 11: 5413-5413
- PubMed: 33110068
- DOI: https://doi.org/10.1038/s41467-020-19231-9
- Primary Citation of Related Structures:
6XE1 - PubMed Abstract:
SARS-CoV-2 is a betacoronavirus virus responsible for the COVID-19 pandemic. Here, we determine the X-ray crystal structure of a potent neutralizing monoclonal antibody, CV30, isolated from a patient infected with SARS-CoV-2, in complex with the receptor binding domain. The structure reveals that CV30 binds to an epitope that overlaps with the human ACE2 receptor binding motif providing a structural basis for its neutralization. CV30 also induces shedding of the S1 subunit, indicating an additional mechanism of neutralization. A germline reversion of CV30 results in a substantial reduction in both binding affinity and neutralization potential indicating the minimal somatic mutation is needed for potently neutralizing antibodies against SARS-CoV-2.
- Fred Hutchinson Cancer Research Center, Vaccines and Infectious Diseases Division, Seattle, WA, USA.
Organizational Affiliation: