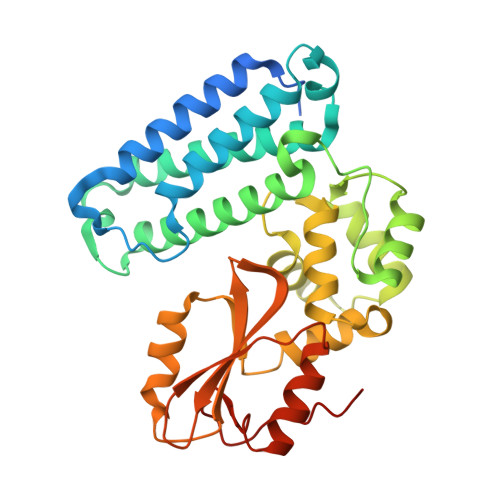
SLAP2 Adaptor Binding Disrupts c-CBL Autoinhibition to Activate Ubiquitin Ligase Function.
Wybenga-Groot, L.E., Tench, A.J., Simpson, C.D., Germain, J.S., Raught, B., Moran, M.F., McGlade, C.J.(2021) J Mol Biology 433: 166880-166880
- PubMed: 33617900
- DOI: https://doi.org/10.1016/j.jmb.2021.166880
- Primary Citation of Related Structures:
6XAR - PubMed Abstract:
CBL is a RING type E3 ubiquitin ligase that functions as a negative regulator of tyrosine kinase signaling and loss of CBL E3 function is implicated in several forms of leukemia. The Src-like adaptor proteins (SLAP/SLAP2) bind to CBL and are required for CBL-dependent downregulation of antigen receptor, cytokine receptor, and receptor tyrosine kinase signaling. Despite the established role of SLAP/SLAP2 in regulating CBL activity, the nature of the interaction and the mechanisms involved are not known. To understand the molecular basis of the interaction between SLAP/SLAP2 and CBL, we solved the crystal structure of CBL tyrosine kinase binding domain (TKBD) in complex with SLAP2. The carboxy-terminal region of SLAP2 adopts an α-helical structure which binds in a cleft between the 4H, EF-hand, and SH2 domains of the TKBD. This SLAP2 binding site is remote from the canonical TKBD phospho-tyrosine peptide binding site but overlaps with a region important for stabilizing CBL in its autoinhibited conformation. In addition, binding of SLAP2 to CBL in vitro activates the ubiquitin ligase function of autoinhibited CBL. Disruption of the CBL/SLAP2 interface through mutagenesis demonstrated a role for this protein-protein interaction in regulation of CBL E3 ligase activity in cells. Our results reveal that SLAP2 binding to a regulatory cleft of the TKBD provides an alternative mechanism for activation of CBL ubiquitin ligase function.
- The Arthur and Sonia Labatt Brain Tumour Research Centre, The Hospital for Sick Children, 555 University Avenue, Toronto, ON M5G 1X8, Canada; Program in Cell Biology, The Hospital for Sick Children, 555 University Avenue, Toronto, ON M5G 1X8, Canada; SPARC BioCentre, The Hospital for Sick Children, 555 University Avenue, Toronto, ON M5G 1X8, Canada. Electronic address: leanne.wybenga-groot@sickkids.ca.
Organizational Affiliation: