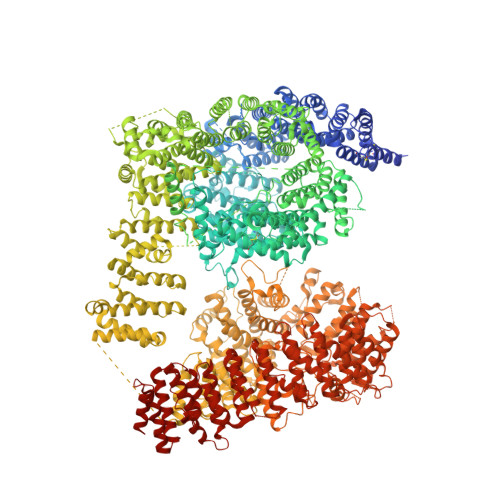
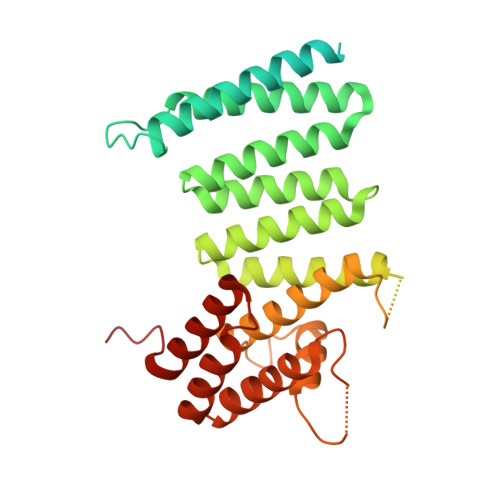
Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1.
Harding, R.J., Deme, J.C., Hevler, J.F., Tamara, S., Lemak, A., Cantle, J.P., Szewczyk, M.M., Begeja, N., Goss, S., Zuo, X., Loppnau, P., Seitova, A., Hutchinson, A., Fan, L., Truant, R., Schapira, M., Carroll, J.B., Heck, A.J.R., Lea, S.M., Arrowsmith, C.H.(2021) Commun Biol 4: 1374-1374
- PubMed: 34880419
- DOI: https://doi.org/10.1038/s42003-021-02895-4
- Primary Citation of Related Structures:
6X9O - PubMed Abstract:
Huntington's disease results from expansion of a glutamine-coding CAG tract in the huntingtin (HTT) gene, producing an aberrantly functioning form of HTT. Both wildtype and disease-state HTT form a hetero-dimer with HAP40 of unknown functional relevance. We demonstrate in vivo and in cell models that HTT and HAP40 cellular abundance are coupled. Integrating data from a 2.6 Å cryo-electron microscopy structure, cross-linking mass spectrometry, small-angle X-ray scattering, and modeling, we provide a near-atomic-level view of HTT, its molecular interaction surfaces and compacted domain architecture, orchestrated by HAP40. Native mass spectrometry reveals a remarkably stable hetero-dimer, potentially explaining the cellular inter-dependence of HTT and HAP40. The exon 1 region of HTT is dynamic but shows greater conformational variety in the polyglutamine expanded mutant than wildtype exon 1. Our data provide a foundation for future functional and drug discovery studies targeting Huntington's disease and illuminate the structural consequences of HTT polyglutamine expansion.
- Structural Genomics Consortium, University of Toronto, Toronto, ON, M5G 1L7, Canada. Rachel.Harding@utoronto.ca.
Organizational Affiliation: