Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Pappalardi, M.B., Keenan, K., Cockerill, M., Kellner, W.A., Stowell, A., Sherk, C., Wong, K., Pathuri, S., Briand, J., Steidel, M., Chapman, P., Groy, A., Wiseman, A.K., McHugh, C.F., Campobasso, N., Graves, A.P., Fairweather, E., Werner, T., Raoof, A., Butlin, R.J., Rueda, L., Horton, J.R., Fosbenner, D.T., Zhang, C., Handler, J.L., Muliaditan, M., Mebrahtu, M., Jaworski, J.P., McNulty, D.E., Burt, C., Eberl, H.C., Taylor, A.N., Ho, T., Merrihew, S., Foley, S.W., Rutkowska, A., Li, M., Romeril, S.P., Goldberg, K., Zhang, X., Kershaw, C.S., Bantscheff, M., Jurewicz, A.J., Minthorn, E., Grandi, P., Patel, M., Benowitz, A.B., Mohammad, H.P., Gilmartin, A.G., Prinjha, R.K., Ogilvie, D., Carpenter, C., Heerding, D., Baylin, S.B., Jones, P.A., Cheng, X., King, B.W., Luengo, J.I., Jordan, A.M., Waddell, I., Kruger, R.G., McCabe, M.T.(2021) Nat Cancer 2: 1002-1017
- PubMed: 34790902
- DOI: https://doi.org/10.3324/haematol.2020.248658
- Primary Citation of Related Structures:
6X9I, 6X9J, 6X9K - PubMed Abstract:
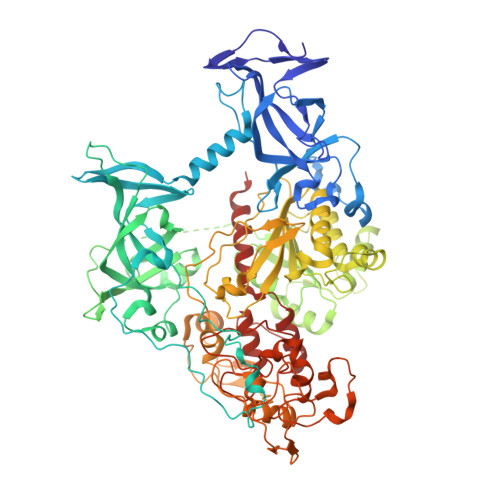
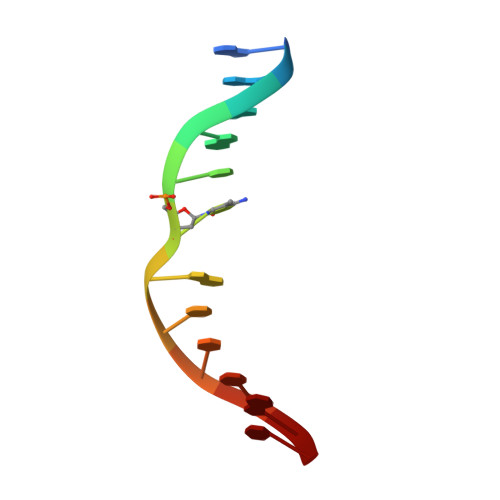
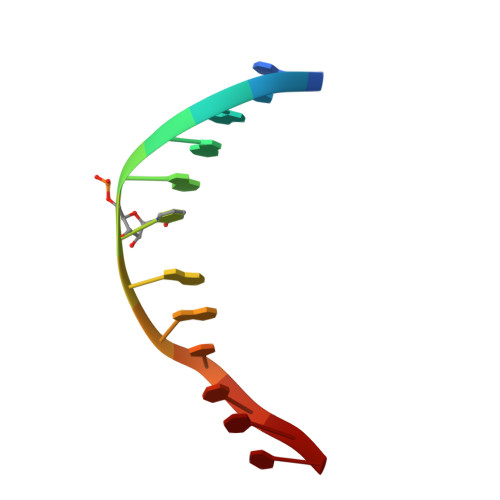
DNA methylation, a key epigenetic driver of transcriptional silencing, is universally dysregulated in cancer. Reversal of DNA methylation by hypomethylating agents, such as the cytidine analogs decitabine or azacytidine, has demonstrated clinical benefit in hematologic malignancies. These nucleoside analogs are incorporated into replicating DNA where they inhibit DNA cytosine methyltransferases DNMT1, DNMT3A and DNMT3B through irreversible covalent interactions. These agents induce notable toxicity to normal blood cells thus limiting their clinical doses. Herein we report the discovery of GSK3685032, a potent first-in-class DNMT1-selective inhibitor that was shown via crystallographic studies to compete with the active-site loop of DNMT1 for penetration into hemi-methylated DNA between two CpG base pairs. GSK3685032 induces robust loss of DNA methylation, transcriptional activation and cancer cell growth inhibition in vitro. Due to improved in vivo tolerability compared with decitabine, GSK3685032 yields superior tumor regression and survival mouse models of acute myeloid leukemia.
- Cancer Epigenetics Research Unit, Oncology, GlaxoSmithKline, Collegeville, PA, USA.
Organizational Affiliation: