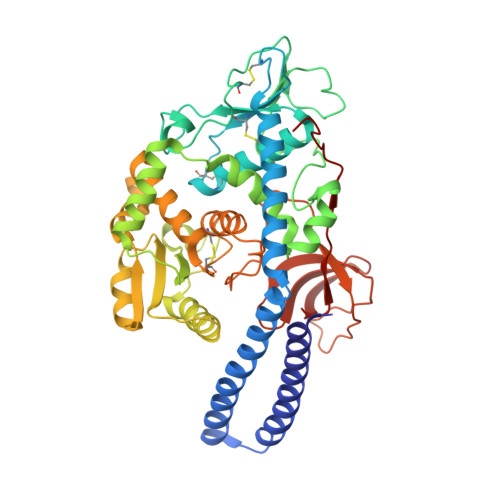
Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
Boruah, B.M., Kadirvelraj, R., Liu, L., Ramiah, A., Li, C., Zong, G., Bosman, G.P., Yang, J.Y., Wang, L.X., Boons, G.J., Wood, Z.A., Moremen, K.W.(2020) J Biological Chem 295: 17027-17045
- PubMed: 33004438
- DOI: https://doi.org/10.1074/jbc.RA120.014625
- Primary Citation of Related Structures:
6X5H, 6X5R, 6X5S, 6X5T, 6X5U - PubMed Abstract:
Mammalian Asn-linked glycans are extensively processed as they transit the secretory pathway to generate diverse glycans on cell surface and secreted glycoproteins. Additional modification of the glycan core by α-1,6-fucose addition to the innermost GlcNAc residue (core fucosylation) is catalyzed by an α-1,6-fucosyltransferase (FUT8). The importance of core fucosylation can be seen in the complex pathological phenotypes of FUT8 null mice, which display defects in cellular signaling, development, and subsequent neonatal lethality. Elevated core fucosylation has also been identified in several human cancers. However, the structural basis for FUT8 substrate specificity remains unknown.Here, using various crystal structures of FUT8 in complex with a donor substrate analog, and with four distinct glycan acceptors, we identify the molecular basis for FUT8 specificity and activity. The ordering of three active site loops corresponds to an increased occupancy for bound GDP, suggesting an induced-fit folding of the donor-binding subsite. Structures of the various acceptor complexes were compared with kinetic data on FUT8 active site mutants and with specificity data from a library of glycan acceptors to reveal how binding site complementarity and steric hindrance can tune substrate affinity. The FUT8 structure was also compared with other known fucosyltransferases to identify conserved and divergent structural features for donor and acceptor recognition and catalysis. These data provide insights into the evolution of modular templates for donor and acceptor recognition among GT-B fold glycosyltransferases in the synthesis of diverse glycan structures in biological systems.
- Complex Carbohydrate Research Center, University of Georgia, Athens, Georgia, USA.
Organizational Affiliation: