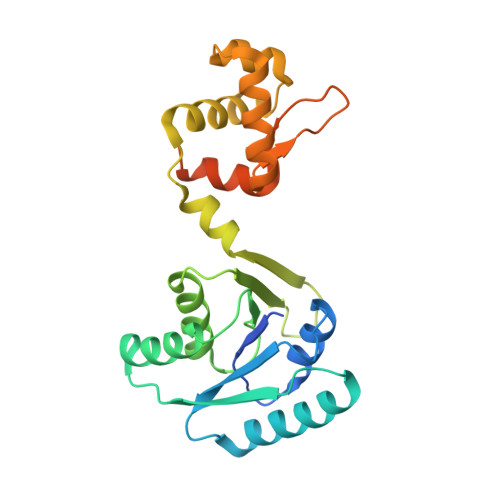
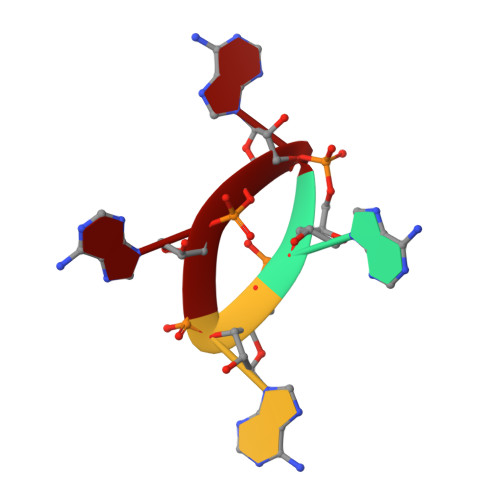
Structural basis of cyclic oligoadenylate binding to the transcription factor Csa3 outlines cross talk between type III and type I CRISPR systems.
Xia, P., Dutta, A., Gupta, K., Batish, M., Parashar, V.(2022) J Biological Chem 298: 101591-101591
- PubMed: 35038453
- DOI: https://doi.org/10.1016/j.jbc.2022.101591
- Primary Citation of Related Structures:
6WXQ - PubMed Abstract:
RNA interference by type III CRISPR systems results in the synthesis of cyclic oligoadenylate (cOA) second messengers, which are known to bind and regulate various CARF domain-containing nuclease receptors. The CARF domain-containing Csa3 family of transcriptional factors associated with the DNA-targeting type I CRISPR systems regulate expression of various CRISPR and DNA repair genes in many prokaryotes. In this study, we extend the known receptor repertoire of cOA messengers to include transcriptional factors by demonstrating specific binding of cyclic tetra-adenylate (cA4) to Saccharolobus solfataricus Csa3 (Csa3 Sso ). Our 2.0-Å resolution X-ray crystal structure of cA4-bound full-length Csa3 Sso reveals the binding of its CARF domain to an elongated conformation of cA4. Using cA4 binding affinity analyses of Csa3 Sso mutants targeting the observed Csa3 Sso •cA4 structural interface, we identified a Csa3-specific cA4 binding motif distinct from a more widely conserved cOA-binding CARF motif. Using a rational surface engineering approach, we increased the cA4 binding affinity of Csa3 Sso up to ∼145-fold over the wildtype, which has potential applications for future second messenger-driven CRISPR gene expression and editing systems. Our in-solution Csa3 Sso structural analysis identified cA4-induced allosteric and asymmetric conformational rearrangement of its C-terminal winged helix-turn-helix effector domains, which could potentially be incompatible to DNA binding. However, specific in vitro binding of the purified Csa3 Sso to its putative promoter (P Cas4a ) was found to be cA4 independent, suggesting a complex mode of Csa3 Sso regulation. Overall, our results support cA4-and Csa3-mediated cross talk between type III and type I CRISPR systems.
- Department of Biological Sciences, University of Delaware, Newark, Delaware, USA.
Organizational Affiliation: