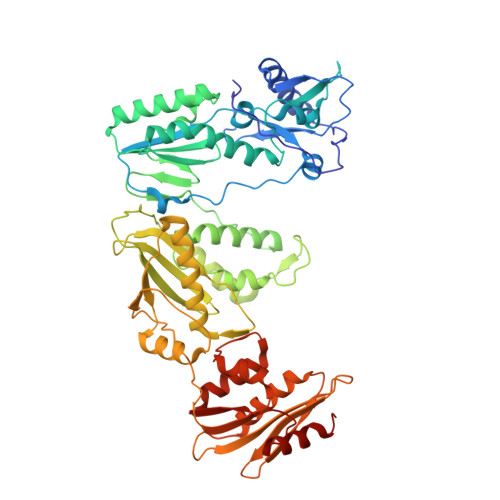
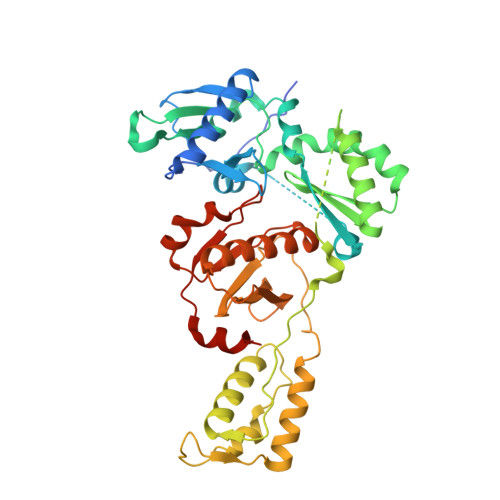
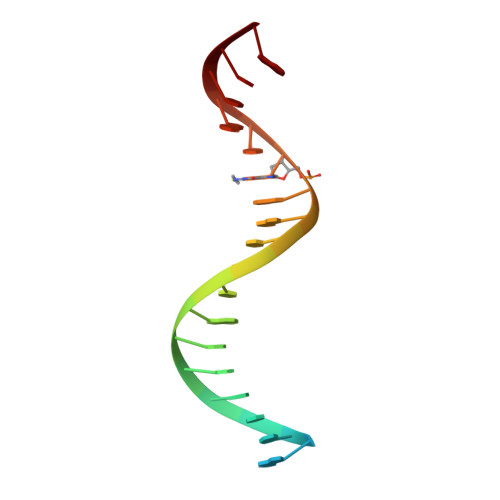
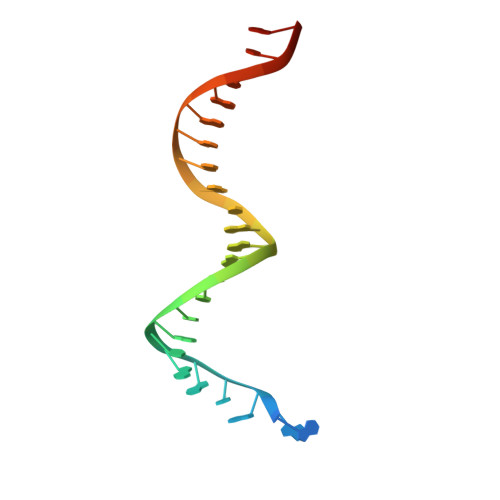
Post-Catalytic Complexes with Emtricitabine or Stavudine and HIV-1 Reverse Transcriptase Reveal New Mechanistic Insights for Nucleotide Incorporation and Drug Resistance.
Bertoletti, N., Chan, A.H., Schinazi, R.F., Anderson, K.S.(2020) Molecules 25
- PubMed: 33096918
- DOI: https://doi.org/10.3390/molecules25204868
- Primary Citation of Related Structures:
6WPF, 6WPH, 6WPJ - PubMed Abstract:
Human immunodeficiency virus 1 (HIV-1) infection is a global health issue since neither a cure nor a vaccine is available. However, the highly active antiretroviral therapy (HAART) has improved the life expectancy for patients with acquired immunodeficiency syndrome (AIDS). Nucleoside reverse transcriptase inhibitors (NRTIs) are in almost all HAART and target reverse transcriptase (RT), an essential enzyme for the virus. Even though NRTIs are highly effective, they have limitations caused by RT resistance. The main mechanisms of RT resistance to NRTIs are discrimination and excision. Understanding the molecular mechanisms for discrimination and excision are essential to develop more potent and selective NRTIs. Using protein X-ray crystallography, we determined the first crystal structure of RT in its post-catalytic state in complex with emtricitabine, (-)FTC or stavudine (d4T). Our structural studies provide the framework for understanding how RT discriminates between NRTIs and natural nucleotides, and for understanding the requirement of (-)FTC to undergo a conformation change for successful incorporation by RT. The crystal structure of RT in post-catalytic complex with d4T provides a "snapshot" for considering the possible mechanism of how RT develops resistance for d4T via excision. The findings reported herein will contribute to the development of next generation NRTIs.
- Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06520-8066, USA.
Organizational Affiliation: