Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Klebsiella pneumoniae in complex with PLP
Stogios, P.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 4-aminobutyrate aminotransferase PuuE | 421 | Klebsiella pneumoniae subsp. pneumoniae SA1 | Mutation(s): 0 Gene Names: puuE, B4U25_33995, BANRA_03059, BL124_00007420, BN49_0292, BVX91_06685, EAO17_12255, EQH49_04735, EQH51_22015, F2X40_16875... EC: 2.6.1.19 | 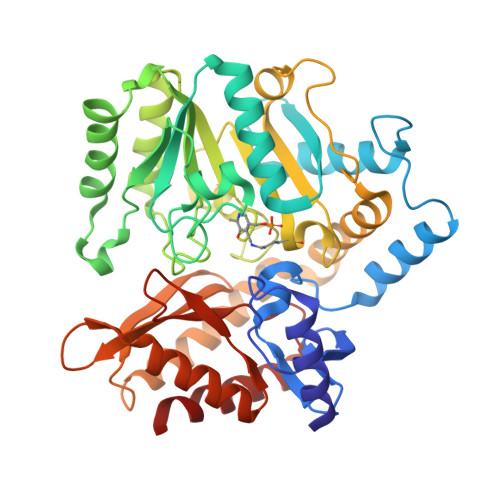 | |
UniProt | |||||
Find proteins for A0A0H3GYJ5 (Klebsiella pneumoniae subsp. pneumoniae (strain HS11286)) Explore A0A0H3GYJ5 Go to UniProtKB: A0A0H3GYJ5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0H3GYJ5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| LLP Query on LLP | A, B, C, D | L-PEPTIDE LINKING | C14 H22 N3 O7 P |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 193.539 | α = 90 |
| b = 63.351 | β = 95.702 |
| c = 128.135 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| PHENIX | phasing |
| PHENIX | model building |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | HHSN272201700060C |