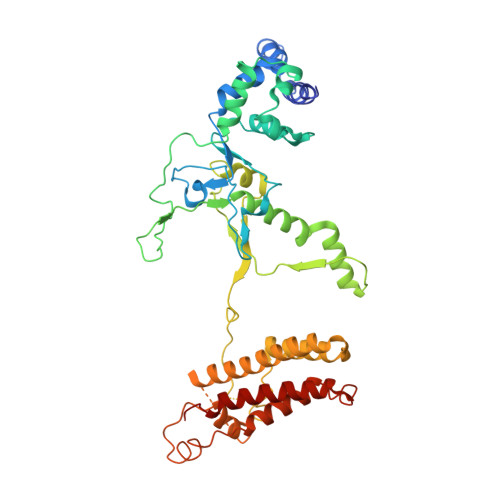
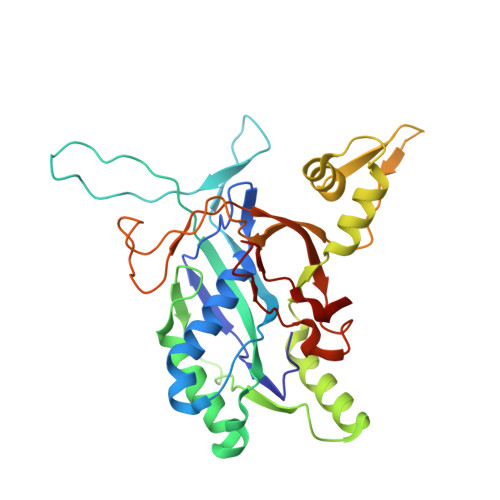
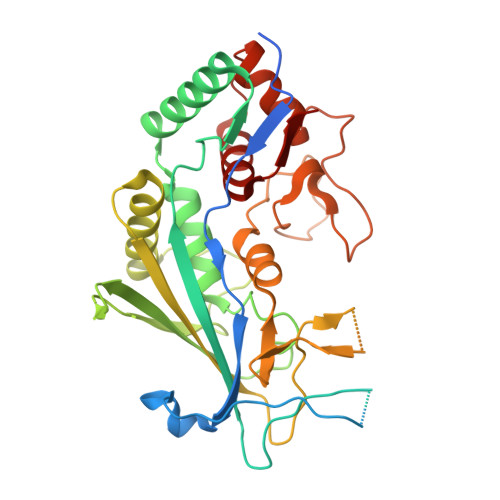
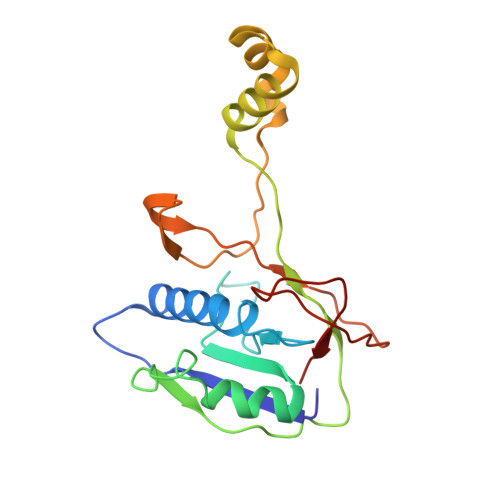
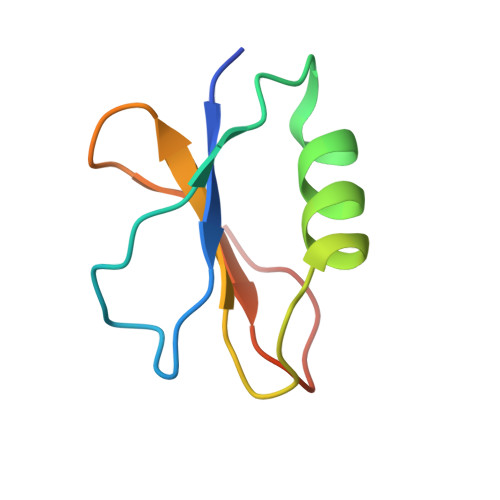
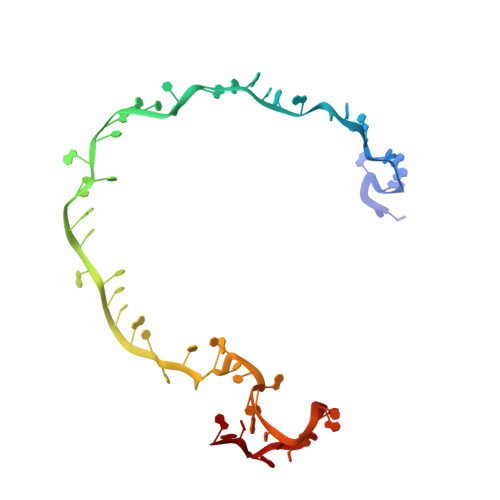
AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex.
Hirschi, M., Lu, W.T., Santiago-Frangos, A., Wilkinson, R., Golden, S.M., Davidson, A.R., Lander, G.C., Wiedenheft, B.(2020) Nat Commun 11: 2730-2730
- PubMed: 32483187
- DOI: https://doi.org/10.1038/s41467-020-16512-1
- Primary Citation of Related Structures:
6W1X, 6WHI - PubMed Abstract:
Bacteria have evolved sophisticated adaptive immune systems, called CRISPR-Cas, that provide sequence-specific protection against phage infection. In turn, phages have evolved a broad spectrum of anti-CRISPRs that suppress these immune systems. Here we report structures of anti-CRISPR protein IF9 (AcrIF9) in complex with the type I-F CRISPR RNA-guided surveillance complex (Csy). In addition to sterically blocking the hybridization of complementary dsDNA to the CRISPR RNA, our results show that AcrIF9 binding also promotes non-sequence-specific engagement with dsDNA, potentially sequestering the complex from target DNA. These findings highlight the versatility of anti-CRISPR mechanisms utilized by phages to suppress CRISPR-mediated immune systems.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA, 92121, USA.
Organizational Affiliation: