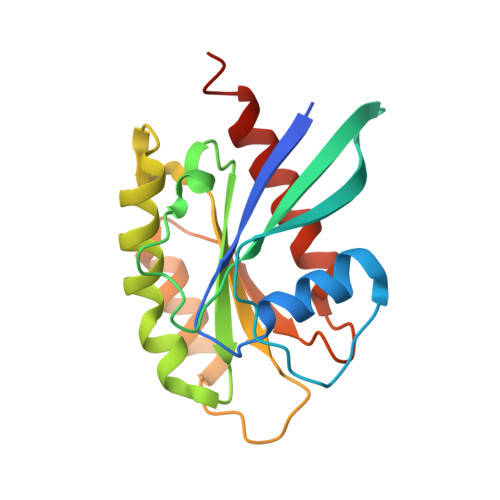
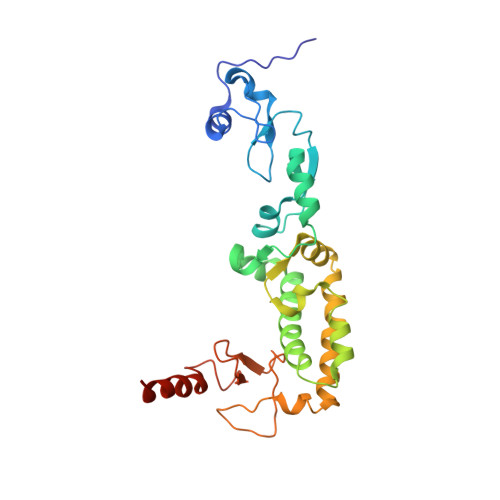
Structural basis for autophagy inhibition by the human Rubicon-Rab7 complex.
Bhargava, H.K., Tabata, K., Byck, J.M., Hamasaki, M., Farrell, D.P., Anishchenko, I., DiMaio, F., Im, Y.J., Yoshimori, T., Hurley, J.H.(2020) Proc Natl Acad Sci U S A 117: 17003-17010
- PubMed: 32632011
- DOI: https://doi.org/10.1073/pnas.2008030117
- Primary Citation of Related Structures:
6WCW - PubMed Abstract:
Rubicon is a potent negative regulator of autophagy and a potential target for autophagy-inducing therapeutics. Rubicon-mediated inhibition of autophagy requires the interaction of the C-terminal Rubicon homology (RH) domain of Rubicon with Rab7-GTP. Here we report the 2.8-Å crystal structure of the Rubicon RH domain in complex with Rab7-GTP. Our structure reveals a fold for the RH domain built around four zinc clusters. The switch regions of Rab7 insert into pockets on the surface of the RH domain in a mode that is distinct from those of other Rab-effector complexes. Rubicon residues at the dimer interface are required for Rubicon and Rab7 to colocalize in living cells. Mutation of Rubicon RH residues in the Rab7-binding site restores efficient autophagic flux in the presence of overexpressed Rubicon, validating the Rubicon RH domain as a promising therapeutic target.
- Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720.
Organizational Affiliation: